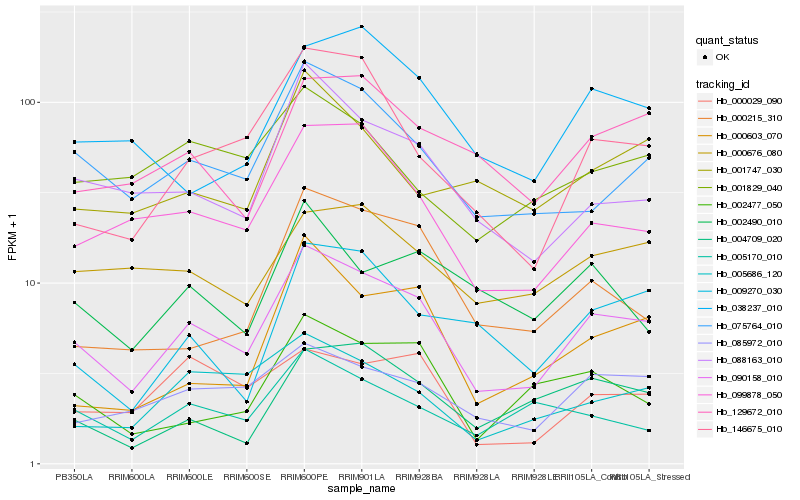
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_090158_010 |
0.0 |
- |
- |
hypothetical protein RCOM_1968400 [Ricinus communis] |
| 2 |
Hb_075764_010 |
0.1269806385 |
- |
- |
PREDICTED: PRA1 family protein A1-like [Jatropha curcas] |
| 3 |
Hb_099878_050 |
0.1336419271 |
- |
- |
PREDICTED: uncharacterized protein LOC105123495 isoform X2 [Populus euphratica] |
| 4 |
Hb_085972_010 |
0.1370081935 |
transcription factor |
TF Family: NAC |
NAC transcription factor 066 [Jatropha curcas] |
| 5 |
Hb_088163_010 |
0.1399176827 |
- |
- |
PREDICTED: glutaminyl-peptide cyclotransferase-like [Jatropha curcas] |
| 6 |
Hb_146675_010 |
0.1447256591 |
- |
- |
PREDICTED: peroxisomal nicotinamide adenine dinucleotide carrier-like isoform X2 [Malus domestica] |
| 7 |
Hb_129672_010 |
0.1472850536 |
- |
- |
PREDICTED: signal peptidase complex subunit 3B [Jatropha curcas] |
| 8 |
Hb_005686_120 |
0.1516840791 |
- |
- |
- |
| 9 |
Hb_002477_050 |
0.1592776002 |
- |
- |
unnamed protein product [Coffea canephora] |
| 10 |
Hb_000676_080 |
0.1592941573 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C17D11.16 [Jatropha curcas] |
| 11 |
Hb_038237_010 |
0.1600967559 |
- |
- |
hypothetical protein POPTR_0003s01110g [Populus trichocarpa] |
| 12 |
Hb_009270_030 |
0.1602648926 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 13 |
Hb_005170_010 |
0.1609990653 |
- |
- |
- |
| 14 |
Hb_000029_090 |
0.1631082877 |
- |
- |
Nicalin precursor, putative [Ricinus communis] |
| 15 |
Hb_004709_020 |
0.163586467 |
- |
- |
PREDICTED: putative threonine aspartase [Jatropha curcas] |
| 16 |
Hb_000603_070 |
0.1635944897 |
- |
- |
- |
| 17 |
Hb_000215_310 |
0.1642563466 |
- |
- |
- |
| 18 |
Hb_001747_030 |
0.1653321893 |
- |
- |
hypothetical protein F383_08210 [Gossypium arboreum] |
| 19 |
Hb_001829_040 |
0.1656242256 |
- |
- |
PREDICTED: protein transport protein SEC13 homolog B [Jatropha curcas] |
| 20 |
Hb_002490_010 |
0.1658931964 |
- |
- |
PREDICTED: plastidic glucose transporter 4 [Jatropha curcas] |