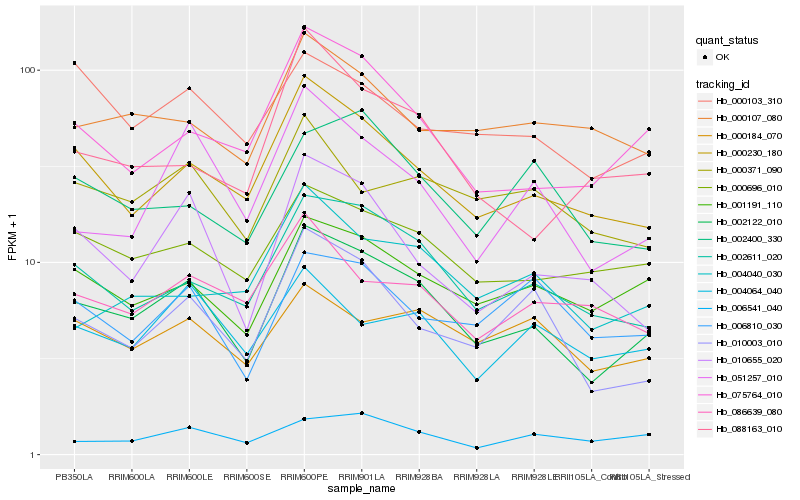
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002122_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_002611_020 |
0.1070380862 |
- |
- |
PREDICTED: alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase-like [Populus euphratica] |
| 3 |
Hb_000230_180 |
0.10930716 |
- |
- |
prolyl 4-hydroxylase alpha subunit, putative [Ricinus communis] |
| 4 |
Hb_075764_010 |
0.1207609911 |
- |
- |
PREDICTED: PRA1 family protein A1-like [Jatropha curcas] |
| 5 |
Hb_001191_110 |
0.1231061782 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 6 |
Hb_000696_010 |
0.1235527247 |
- |
- |
RING-H2 finger protein ATL4M, putative [Ricinus communis] |
| 7 |
Hb_010003_010 |
0.1304594545 |
- |
- |
PREDICTED: uncharacterized protein LOC105630174 isoform X2 [Jatropha curcas] |
| 8 |
Hb_010655_020 |
0.1361319547 |
- |
- |
hypothetical protein CICLE_v10032410mg [Citrus clementina] |
| 9 |
Hb_051257_010 |
0.1379935002 |
- |
- |
- |
| 10 |
Hb_006541_040 |
0.1460203699 |
- |
- |
PREDICTED: diphthamide biosynthesis protein 2 [Jatropha curcas] |
| 11 |
Hb_000371_090 |
0.1483147997 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit-like [Jatropha curcas] |
| 12 |
Hb_004064_040 |
0.1493010902 |
- |
- |
PREDICTED: Down syndrome critical region protein 3 homolog isoform X3 [Jatropha curcas] |
| 13 |
Hb_088163_010 |
0.1493020266 |
- |
- |
PREDICTED: glutaminyl-peptide cyclotransferase-like [Jatropha curcas] |
| 14 |
Hb_000184_070 |
0.1497629017 |
- |
- |
PREDICTED: uncharacterized protein LOC105641537 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000107_080 |
0.14994543 |
- |
- |
PREDICTED: uncharacterized protein LOC105641009 [Jatropha curcas] |
| 16 |
Hb_004040_030 |
0.1513644778 |
- |
- |
PREDICTED: probable inactive purple acid phosphatase 28 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000103_310 |
0.1519751158 |
- |
- |
PREDICTED: transmembrane emp24 domain-containing protein p24beta3 [Jatropha curcas] |
| 18 |
Hb_002400_330 |
0.1521984071 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |
| 19 |
Hb_006810_030 |
0.1527950225 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 20 |
Hb_086639_080 |
0.1531007529 |
- |
- |
conserved hypothetical protein [Ricinus communis] |