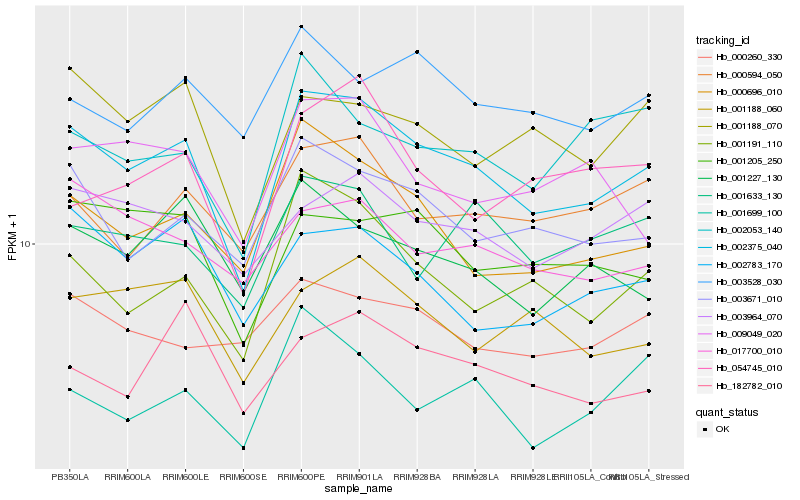
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002375_040 |
0.0 |
- |
- |
PREDICTED: 15 kDa selenoprotein [Jatropha curcas] |
| 2 |
Hb_003671_010 |
0.0879419904 |
- |
- |
PREDICTED: protein transport protein SFT2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000696_010 |
0.0890186441 |
- |
- |
RING-H2 finger protein ATL4M, putative [Ricinus communis] |
| 4 |
Hb_001191_110 |
0.0897488472 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 5 |
Hb_001205_250 |
0.0971342328 |
- |
- |
PREDICTED: uncharacterized protein LOC105643856 [Jatropha curcas] |
| 6 |
Hb_003964_070 |
0.1005181825 |
- |
- |
hypothetical protein JCGZ_15695 [Jatropha curcas] |
| 7 |
Hb_000594_050 |
0.1010064219 |
- |
- |
PREDICTED: pH-response regulator protein palA/RIM20 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001188_070 |
0.1019435647 |
- |
- |
PREDICTED: protein disulfide-isomerase 5-1 [Jatropha curcas] |
| 9 |
Hb_001188_060 |
0.1029869301 |
- |
- |
- |
| 10 |
Hb_001227_130 |
0.1032203562 |
- |
- |
PREDICTED: delta(14)-sterol reductase [Jatropha curcas] |
| 11 |
Hb_002053_140 |
0.1043463992 |
- |
- |
PREDICTED: coatomer subunit epsilon-1 [Jatropha curcas] |
| 12 |
Hb_009049_020 |
0.1052171769 |
- |
- |
PREDICTED: 2-dehydro-3-deoxyphosphooctonate aldolase 1-like [Jatropha curcas] |
| 13 |
Hb_001633_130 |
0.1052823145 |
- |
- |
PREDICTED: uncharacterized protein LOC105630200 [Jatropha curcas] |
| 14 |
Hb_002783_170 |
0.1055054611 |
- |
- |
PREDICTED: BRCA1-A complex subunit BRE isoform X2 [Jatropha curcas] |
| 15 |
Hb_017700_010 |
0.1057599518 |
- |
- |
PREDICTED: probable protein S-acyltransferase 15 [Jatropha curcas] |
| 16 |
Hb_003528_030 |
0.1074962601 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit L-like [Jatropha curcas] |
| 17 |
Hb_001699_100 |
0.1085458584 |
- |
- |
PREDICTED: calmodulin-lysine N-methyltransferase [Jatropha curcas] |
| 18 |
Hb_182782_010 |
0.1087626227 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_054745_010 |
0.1102713207 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000260_330 |
0.1103074308 |
- |
- |
PREDICTED: uncharacterized protein LOC103438625 [Malus domestica] |