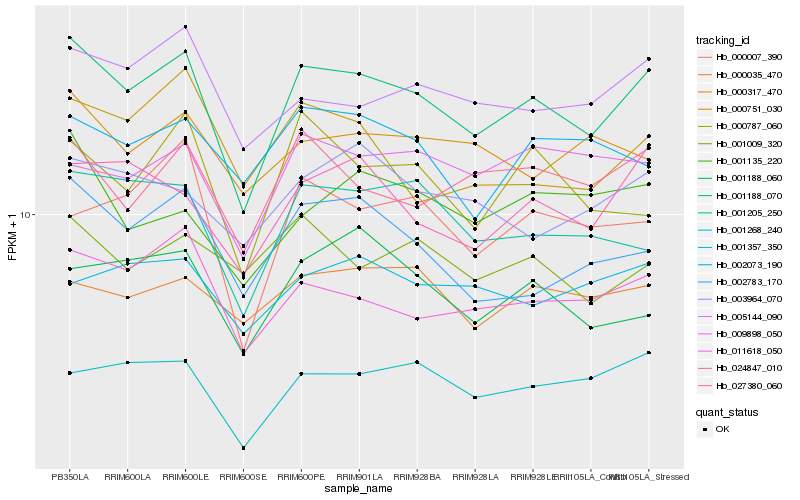
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001188_070 |
0.0 |
- |
- |
PREDICTED: protein disulfide-isomerase 5-1 [Jatropha curcas] |
| 2 |
Hb_011618_050 |
0.0770189281 |
- |
- |
PREDICTED: uncharacterized protein LOC105634948 [Jatropha curcas] |
| 3 |
Hb_001268_240 |
0.0815581339 |
- |
- |
PREDICTED: uncharacterized protein LOC105630174 isoform X2 [Jatropha curcas] |
| 4 |
Hb_024847_010 |
0.0842094925 |
- |
- |
UPF0586 C9orf41-like protein [Medicago truncatula] |
| 5 |
Hb_001205_250 |
0.0844977445 |
- |
- |
PREDICTED: uncharacterized protein LOC105643856 [Jatropha curcas] |
| 6 |
Hb_005144_090 |
0.084882519 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 24 homolog 1-like [Citrus sinensis] |
| 7 |
Hb_002783_170 |
0.0854980001 |
- |
- |
PREDICTED: BRCA1-A complex subunit BRE isoform X2 [Jatropha curcas] |
| 8 |
Hb_027380_060 |
0.0864776015 |
- |
- |
PREDICTED: putative ALA-interacting subunit 2 [Jatropha curcas] |
| 9 |
Hb_000787_060 |
0.0943368649 |
- |
- |
thioredoxin domain-containing protein, putative [Ricinus communis] |
| 10 |
Hb_009898_050 |
0.0946629638 |
- |
- |
PREDICTED: beta-taxilin [Jatropha curcas] |
| 11 |
Hb_001188_060 |
0.0950640593 |
- |
- |
- |
| 12 |
Hb_003964_070 |
0.0957454832 |
- |
- |
hypothetical protein JCGZ_15695 [Jatropha curcas] |
| 13 |
Hb_000007_390 |
0.0958762448 |
- |
- |
hypothetical protein JCGZ_18250 [Jatropha curcas] |
| 14 |
Hb_000035_470 |
0.0959195317 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001135_220 |
0.0962415738 |
- |
- |
FGFR1 oncogene partner, putative [Ricinus communis] |
| 16 |
Hb_000317_470 |
0.0976234305 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 11 [Jatropha curcas] |
| 17 |
Hb_002073_190 |
0.0982538831 |
- |
- |
PREDICTED: uncharacterized protein LOC105649812 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000751_030 |
0.0986363704 |
- |
- |
Vesicle transport protein GOT1B, putative [Ricinus communis] |
| 19 |
Hb_001357_350 |
0.0994294845 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001009_320 |
0.0996578558 |
- |
- |
PREDICTED: uncharacterized protein LOC105640250 [Jatropha curcas] |