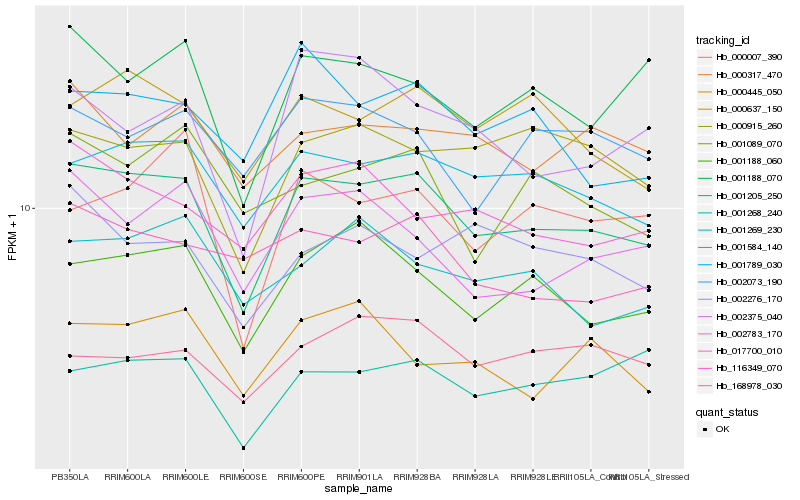
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001205_250 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643856 [Jatropha curcas] |
| 2 |
Hb_001584_140 |
0.069733717 |
- |
- |
PREDICTED: uncharacterized protein LOC105647409 [Jatropha curcas] |
| 3 |
Hb_001188_060 |
0.0749531058 |
- |
- |
- |
| 4 |
Hb_000637_150 |
0.0828991116 |
- |
- |
Protein CASP, putative [Ricinus communis] |
| 5 |
Hb_001188_070 |
0.0844977445 |
- |
- |
PREDICTED: protein disulfide-isomerase 5-1 [Jatropha curcas] |
| 6 |
Hb_000915_260 |
0.0883809452 |
- |
- |
PREDICTED: uncharacterized protein LOC105628514 isoform X1 [Jatropha curcas] |
| 7 |
Hb_017700_010 |
0.0899622778 |
- |
- |
PREDICTED: probable protein S-acyltransferase 15 [Jatropha curcas] |
| 8 |
Hb_002783_170 |
0.0914252539 |
- |
- |
PREDICTED: BRCA1-A complex subunit BRE isoform X2 [Jatropha curcas] |
| 9 |
Hb_000317_470 |
0.0920887859 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 11 [Jatropha curcas] |
| 10 |
Hb_001789_030 |
0.0937418483 |
- |
- |
PREDICTED: signal peptide peptidase-like 1 [Jatropha curcas] |
| 11 |
Hb_001269_230 |
0.0937947395 |
- |
- |
PREDICTED: tRNA-splicing endonuclease subunit Sen54 [Jatropha curcas] |
| 12 |
Hb_001268_240 |
0.0944651749 |
- |
- |
PREDICTED: uncharacterized protein LOC105630174 isoform X2 [Jatropha curcas] |
| 13 |
Hb_002073_190 |
0.0951192916 |
- |
- |
PREDICTED: uncharacterized protein LOC105649812 isoform X1 [Jatropha curcas] |
| 14 |
Hb_116349_070 |
0.0958378549 |
- |
- |
hypothetical protein JCGZ_21753 [Jatropha curcas] |
| 15 |
Hb_000007_390 |
0.0961659769 |
- |
- |
hypothetical protein JCGZ_18250 [Jatropha curcas] |
| 16 |
Hb_002375_040 |
0.0971342328 |
- |
- |
PREDICTED: 15 kDa selenoprotein [Jatropha curcas] |
| 17 |
Hb_001089_070 |
0.0974787919 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 3 [Jatropha curcas] |
| 18 |
Hb_168978_030 |
0.0975216916 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 8 [Jatropha curcas] |
| 19 |
Hb_000445_050 |
0.0978987441 |
- |
- |
PREDICTED: DNA topoisomerase 2-binding protein 1 [Jatropha curcas] |
| 20 |
Hb_002276_170 |
0.0979539318 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |