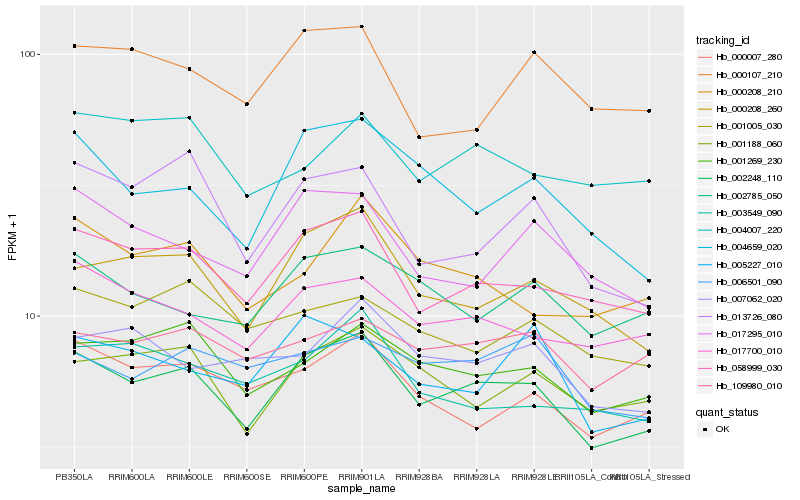
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002248_110 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s41050g [Populus trichocarpa] |
| 2 |
Hb_058999_030 |
0.0629521013 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 3 |
Hb_000208_260 |
0.0769220834 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4 [Jatropha curcas] |
| 4 |
Hb_001269_230 |
0.0817478763 |
- |
- |
PREDICTED: tRNA-splicing endonuclease subunit Sen54 [Jatropha curcas] |
| 5 |
Hb_017700_010 |
0.0827809713 |
- |
- |
PREDICTED: probable protein S-acyltransferase 15 [Jatropha curcas] |
| 6 |
Hb_001188_060 |
0.0853246249 |
- |
- |
- |
| 7 |
Hb_004659_020 |
0.0866622497 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 8 |
Hb_000007_280 |
0.0883021836 |
- |
- |
hypothetical protein VITISV_000066 [Vitis vinifera] |
| 9 |
Hb_017295_010 |
0.0902356747 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g06890 [Jatropha curcas] |
| 10 |
Hb_002785_050 |
0.0920394919 |
- |
- |
PREDICTED: adenylosuccinate lyase-like [Jatropha curcas] |
| 11 |
Hb_013726_080 |
0.0937516384 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 12 |
Hb_000208_210 |
0.0942270445 |
- |
- |
PREDICTED: RNA-binding protein Nova-2 [Jatropha curcas] |
| 13 |
Hb_109980_010 |
0.0953643514 |
- |
- |
PREDICTED: uncharacterized protein LOC105647182 isoform X1 [Jatropha curcas] |
| 14 |
Hb_005227_010 |
0.0977858922 |
- |
- |
PREDICTED: uncharacterized protein LOC105633453 isoform X1 [Jatropha curcas] |
| 15 |
Hb_007062_020 |
0.0978110856 |
- |
- |
gcn4-complementing protein, putative [Ricinus communis] |
| 16 |
Hb_006501_090 |
0.0978406255 |
- |
- |
PREDICTED: golgin candidate 1 [Jatropha curcas] |
| 17 |
Hb_001005_030 |
0.0980572655 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 18 |
Hb_004007_220 |
0.0985518066 |
- |
- |
PREDICTED: casein kinase II subunit alpha [Jatropha curcas] |
| 19 |
Hb_003549_090 |
0.0989594973 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 6 [Jatropha curcas] |
| 20 |
Hb_000107_210 |
0.0992474104 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 2 [Jatropha curcas] |