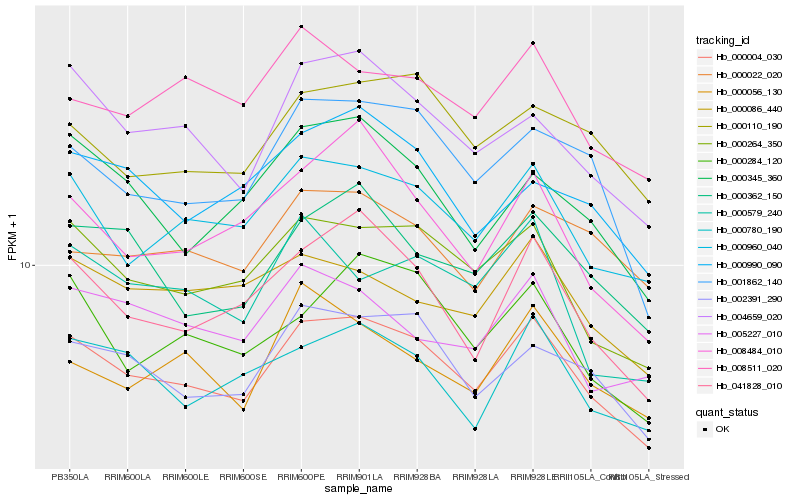
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000004_030 |
0.0 |
- |
- |
PREDICTED: Niemann-Pick C1 protein [Jatropha curcas] |
| 2 |
Hb_000264_350 |
0.0619958211 |
- |
- |
PREDICTED: uncharacterized protein LOC105649541 [Jatropha curcas] |
| 3 |
Hb_000960_040 |
0.065301764 |
- |
- |
PREDICTED: ERAD-associated E3 ubiquitin-protein ligase HRD1B-like [Vitis vinifera] |
| 4 |
Hb_041828_010 |
0.0706676473 |
- |
- |
PREDICTED: alpha-mannosidase [Jatropha curcas] |
| 5 |
Hb_002391_290 |
0.0727398259 |
- |
- |
PREDICTED: bifunctional nuclease 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004659_020 |
0.075814534 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 7 |
Hb_001862_140 |
0.0760378101 |
- |
- |
natural resistance-associated macrophage protein, putative [Ricinus communis] |
| 8 |
Hb_000362_150 |
0.080556674 |
- |
- |
PREDICTED: homogentisate 1,2-dioxygenase [Jatropha curcas] |
| 9 |
Hb_000780_190 |
0.082569706 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 2 [Jatropha curcas] |
| 10 |
Hb_000284_120 |
0.0860124684 |
- |
- |
PREDICTED: uncharacterized protein LOC105629656 [Jatropha curcas] |
| 11 |
Hb_000345_360 |
0.0862313123 |
- |
- |
PREDICTED: protein AUXIN RESPONSE 4 [Jatropha curcas] |
| 12 |
Hb_000110_190 |
0.08851802 |
- |
- |
hypothetical protein B456_013G081300 [Gossypium raimondii] |
| 13 |
Hb_008484_010 |
0.0899420811 |
- |
- |
hypothetical protein B456_008G050700 [Gossypium raimondii] |
| 14 |
Hb_000086_440 |
0.0903406296 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 15 |
Hb_008511_020 |
0.0916973701 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 16 |
Hb_000990_090 |
0.0921778975 |
- |
- |
PREDICTED: ER membrane protein complex subunit 1 [Jatropha curcas] |
| 17 |
Hb_000579_240 |
0.0950249414 |
- |
- |
phosphatidylinositol transporter, putative [Ricinus communis] |
| 18 |
Hb_005227_010 |
0.0950682726 |
- |
- |
PREDICTED: uncharacterized protein LOC105633453 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000056_130 |
0.0954235088 |
- |
- |
Sucrose Transporter 2C [Hevea brasiliensis subsp. brasiliensis] |
| 20 |
Hb_000022_020 |
0.0967902768 |
- |
- |
hypothetical protein CICLE_v10020565mg [Citrus clementina] |