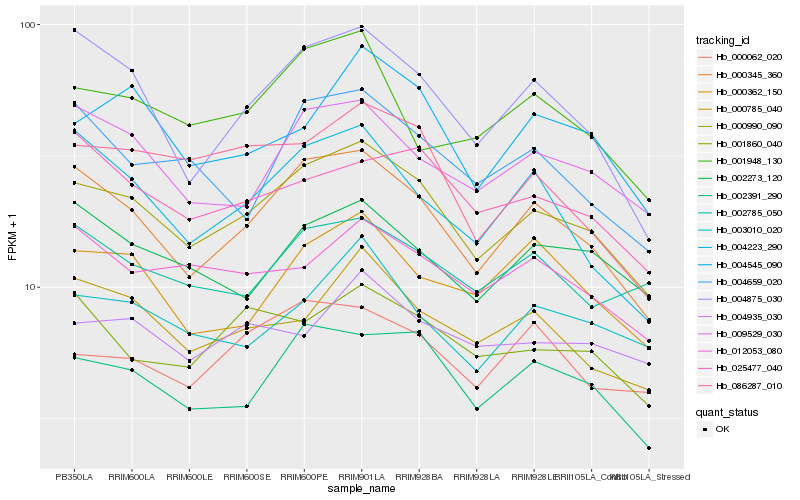
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000990_090 |
0.0 |
- |
- |
PREDICTED: ER membrane protein complex subunit 1 [Jatropha curcas] |
| 2 |
Hb_000345_360 |
0.0507834266 |
- |
- |
PREDICTED: protein AUXIN RESPONSE 4 [Jatropha curcas] |
| 3 |
Hb_002391_290 |
0.0666452886 |
- |
- |
PREDICTED: bifunctional nuclease 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_009529_030 |
0.0783009281 |
- |
- |
PREDICTED: transmembrane protein 120 homolog [Jatropha curcas] |
| 5 |
Hb_000785_040 |
0.0812812518 |
- |
- |
PREDICTED: uncharacterized protein LOC105628919 [Jatropha curcas] |
| 6 |
Hb_004875_030 |
0.0815444655 |
- |
- |
PREDICTED: signal peptide peptidase-like 2 [Jatropha curcas] |
| 7 |
Hb_004545_090 |
0.0836310298 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003010_020 |
0.085295206 |
- |
- |
PREDICTED: probable inactive heme oxygenase 2, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_086287_010 |
0.0861108239 |
- |
- |
PREDICTED: aminoacylase-1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000062_020 |
0.0863446078 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 11 |
Hb_001860_040 |
0.0864948154 |
- |
- |
PREDICTED: cleavage stimulation factor subunit 77 isoform X4 [Jatropha curcas] |
| 12 |
Hb_012053_080 |
0.0876908735 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 13 |
Hb_002273_120 |
0.0883055484 |
- |
- |
PREDICTED: probable apyrase 6 [Jatropha curcas] |
| 14 |
Hb_001948_130 |
0.0893768873 |
- |
- |
PREDICTED: uncharacterized protein LOC105638579 [Jatropha curcas] |
| 15 |
Hb_004659_020 |
0.0898969324 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 16 |
Hb_000362_150 |
0.0905714421 |
- |
- |
PREDICTED: homogentisate 1,2-dioxygenase [Jatropha curcas] |
| 17 |
Hb_002785_050 |
0.0909999723 |
- |
- |
PREDICTED: adenylosuccinate lyase-like [Jatropha curcas] |
| 18 |
Hb_004223_290 |
0.0910041476 |
- |
- |
PREDICTED: protein IQ-DOMAIN 32 [Jatropha curcas] |
| 19 |
Hb_025477_040 |
0.0911127466 |
- |
- |
PREDICTED: post-GPI attachment to proteins factor 3 [Jatropha curcas] |
| 20 |
Hb_004935_030 |
0.0914785505 |
- |
- |
PREDICTED: uncharacterized protein LOC102628125 [Citrus sinensis] |