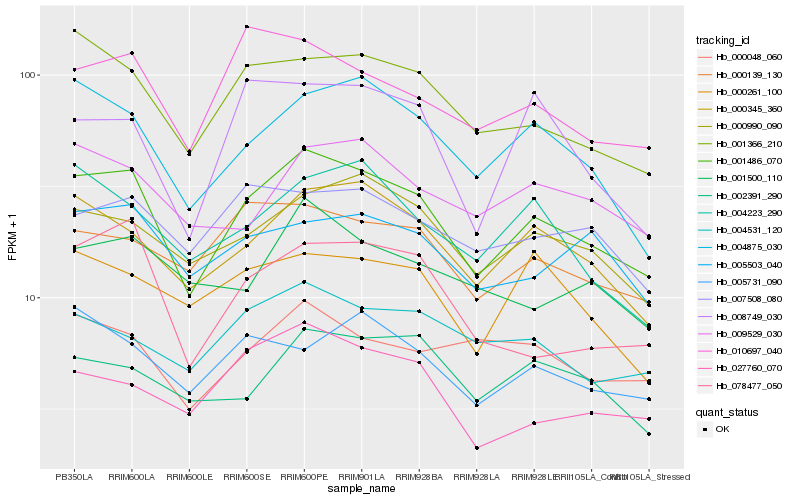
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001486_070 |
0.0 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 7 [Jatropha curcas] |
| 2 |
Hb_000345_360 |
0.0863476494 |
- |
- |
PREDICTED: protein AUXIN RESPONSE 4 [Jatropha curcas] |
| 3 |
Hb_004875_030 |
0.0923928615 |
- |
- |
PREDICTED: signal peptide peptidase-like 2 [Jatropha curcas] |
| 4 |
Hb_001366_210 |
0.0928000681 |
- |
- |
cathepsin B, putative [Ricinus communis] |
| 5 |
Hb_000990_090 |
0.0982119033 |
- |
- |
PREDICTED: ER membrane protein complex subunit 1 [Jatropha curcas] |
| 6 |
Hb_000139_130 |
0.107672309 |
- |
- |
PREDICTED: chloride channel protein CLC-d isoform X1 [Jatropha curcas] |
| 7 |
Hb_078477_050 |
0.1077498072 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105648332 isoform X1 [Jatropha curcas] |
| 8 |
Hb_010697_040 |
0.1085095649 |
- |
- |
PREDICTED: protein GPR107 isoform X1 [Jatropha curcas] |
| 9 |
Hb_005503_040 |
0.1114942691 |
- |
- |
PREDICTED: AMP deaminase [Jatropha curcas] |
| 10 |
Hb_004223_290 |
0.1117098712 |
- |
- |
PREDICTED: protein IQ-DOMAIN 32 [Jatropha curcas] |
| 11 |
Hb_004531_120 |
0.1122284124 |
- |
- |
PREDICTED: uncharacterized protein LOC105635025 [Jatropha curcas] |
| 12 |
Hb_009529_030 |
0.1134933939 |
- |
- |
PREDICTED: transmembrane protein 120 homolog [Jatropha curcas] |
| 13 |
Hb_005731_090 |
0.1137387758 |
- |
- |
PREDICTED: uncharacterized protein LOC105630164 [Jatropha curcas] |
| 14 |
Hb_007508_080 |
0.1142621577 |
- |
- |
PREDICTED: presenilin-like protein At2g29900 [Jatropha curcas] |
| 15 |
Hb_027760_070 |
0.1156774908 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02-like [Jatropha curcas] |
| 16 |
Hb_000048_060 |
0.1176395792 |
- |
- |
PREDICTED: probable apyrase 7 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000261_100 |
0.1181152603 |
- |
- |
PREDICTED: beta-galactosidase 9 isoform X2 [Jatropha curcas] |
| 18 |
Hb_008749_030 |
0.118853388 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 19 |
Hb_001500_110 |
0.1191806092 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_002391_290 |
0.1193230953 |
- |
- |
PREDICTED: bifunctional nuclease 2 isoform X1 [Jatropha curcas] |