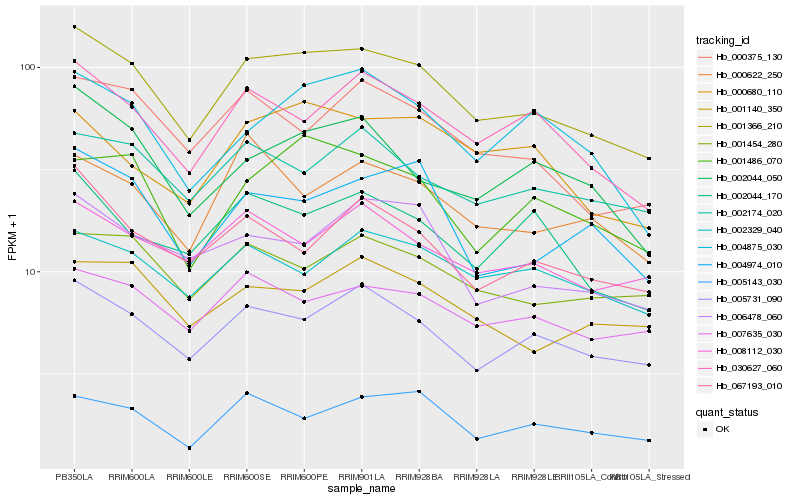
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001366_210 |
0.0 |
- |
- |
cathepsin B, putative [Ricinus communis] |
| 2 |
Hb_005731_090 |
0.0690234161 |
- |
- |
PREDICTED: uncharacterized protein LOC105630164 [Jatropha curcas] |
| 3 |
Hb_030627_060 |
0.0810752766 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 4 |
Hb_007635_030 |
0.0864927849 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g77360, mitochondrial-like [Jatropha curcas] |
| 5 |
Hb_000375_130 |
0.0892708969 |
- |
- |
Leucine-rich repeat transmembrane protein kinase family protein isoform 1 [Theobroma cacao] |
| 6 |
Hb_001454_280 |
0.0902266959 |
- |
- |
PREDICTED: chaperone protein dnaJ 49 [Jatropha curcas] |
| 7 |
Hb_002174_020 |
0.0919085662 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 8 |
Hb_008112_030 |
0.0919113165 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001486_070 |
0.0928000681 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 7 [Jatropha curcas] |
| 10 |
Hb_004974_010 |
0.0938315368 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880-like [Prunus mume] |
| 11 |
Hb_006478_060 |
0.0944868022 |
- |
- |
PREDICTED: homocysteine S-methyltransferase 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002044_050 |
0.0945408883 |
- |
- |
- |
| 13 |
Hb_001140_350 |
0.0947159062 |
- |
- |
PREDICTED: uncharacterized protein LOC105648465 [Jatropha curcas] |
| 14 |
Hb_002329_040 |
0.0947397031 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 15 |
Hb_004875_030 |
0.0948959206 |
- |
- |
PREDICTED: signal peptide peptidase-like 2 [Jatropha curcas] |
| 16 |
Hb_005143_030 |
0.0951623071 |
- |
- |
PREDICTED: uncharacterized protein LOC105638843 [Jatropha curcas] |
| 17 |
Hb_002044_170 |
0.0951818995 |
- |
- |
hypothetical protein POPTR_0014s17160g [Populus trichocarpa] |
| 18 |
Hb_067193_010 |
0.0958561221 |
- |
- |
- |
| 19 |
Hb_000680_110 |
0.0964461306 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 20 |
Hb_000622_250 |
0.0985313393 |
- |
- |
atpob1, putative [Ricinus communis] |