| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
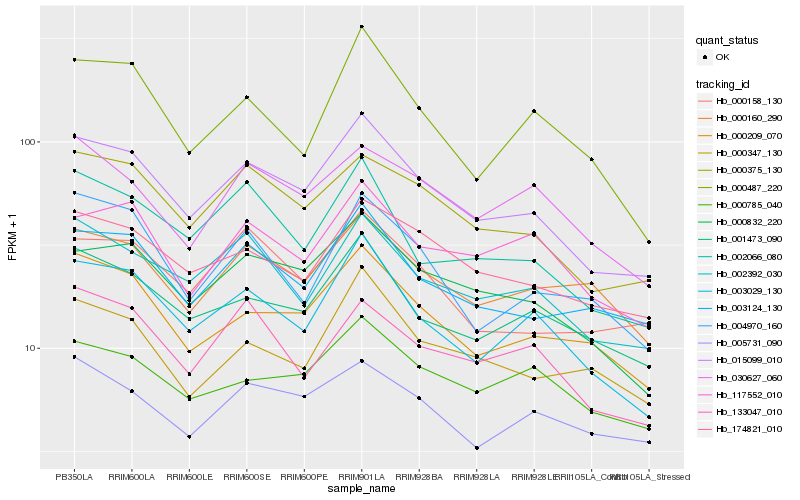
Hb_015099_010 |
0.0 |
- |
- |
PREDICTED: probable glucan 1,3-alpha-glucosidase [Jatropha curcas] |
| 2 |
Hb_000375_130 |
0.0593904482 |
- |
- |
Leucine-rich repeat transmembrane protein kinase family protein isoform 1 [Theobroma cacao] |
| 3 |
Hb_000832_220 |
0.0613300991 |
- |
- |
PREDICTED: uncharacterized protein LOC105641576 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003029_130 |
0.0632813329 |
- |
- |
carboxypeptidase regulatory region-containingprotein, putative [Ricinus communis] |
| 5 |
Hb_002392_030 |
0.0712355256 |
- |
- |
WD-40 repeat family protein / small nuclear ribonucleoprotein Prp4p-related [Theobroma cacao] |
| 6 |
Hb_003124_130 |
0.0721139201 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000209_070 |
0.072643049 |
- |
- |
PREDICTED: chloride channel protein CLC-f isoform X1 [Jatropha curcas] |
| 8 |
Hb_001473_090 |
0.0754921019 |
- |
- |
PREDICTED: uncharacterized protein LOC105650374 [Jatropha curcas] |
| 9 |
Hb_174821_010 |
0.0762612601 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 14 [Jatropha curcas] |
| 10 |
Hb_002066_080 |
0.0795761777 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 11 |
Hb_000158_130 |
0.084171501 |
- |
- |
PREDICTED: uncharacterized protein LOC105647950 isoform X1 [Jatropha curcas] |
| 12 |
Hb_117552_010 |
0.0870310007 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_000160_290 |
0.0890642071 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_133047_010 |
0.0899408432 |
- |
- |
xanthine dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_000785_040 |
0.0912649566 |
- |
- |
PREDICTED: uncharacterized protein LOC105628919 [Jatropha curcas] |
| 16 |
Hb_000347_130 |
0.0913645364 |
- |
- |
PREDICTED: nuclear pore complex protein NUP107 [Jatropha curcas] |
| 17 |
Hb_030627_060 |
0.0920765016 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 18 |
Hb_000487_220 |
0.0930827486 |
- |
- |
eukaryotic translation elongation factor, putative [Ricinus communis] |
| 19 |
Hb_005731_090 |
0.0932842841 |
- |
- |
PREDICTED: uncharacterized protein LOC105630164 [Jatropha curcas] |
| 20 |
Hb_004970_160 |
0.0934588087 |
- |
- |
PREDICTED: THO complex subunit 5B-like isoform X2 [Jatropha curcas] |