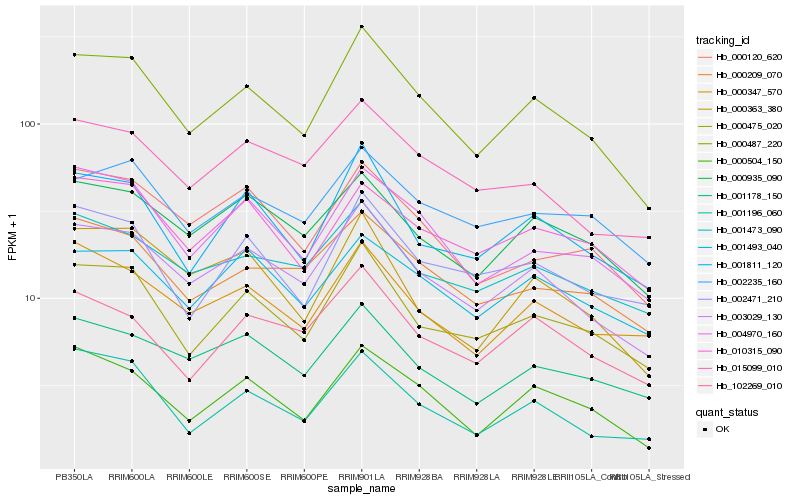
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000487_220 |
0.0 |
- |
- |
eukaryotic translation elongation factor, putative [Ricinus communis] |
| 2 |
Hb_003029_130 |
0.0481077743 |
- |
- |
carboxypeptidase regulatory region-containingprotein, putative [Ricinus communis] |
| 3 |
Hb_000475_020 |
0.0675990858 |
- |
- |
PREDICTED: dehydrogenase/reductase SDR family member on chromosome X homolog isoform X2 [Jatropha curcas] |
| 4 |
Hb_000504_150 |
0.0689334452 |
- |
- |
PREDICTED: nuclear pore complex protein GP210 [Jatropha curcas] |
| 5 |
Hb_001811_120 |
0.0748508215 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004970_160 |
0.0771861024 |
- |
- |
PREDICTED: THO complex subunit 5B-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_000209_070 |
0.0894321606 |
- |
- |
PREDICTED: chloride channel protein CLC-f isoform X1 [Jatropha curcas] |
| 8 |
Hb_000935_090 |
0.0902219774 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 9 |
Hb_001178_150 |
0.0921979557 |
- |
- |
PREDICTED: uncharacterized protein LOC105635484 [Jatropha curcas] |
| 10 |
Hb_001196_060 |
0.0922443461 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 11 |
Hb_002471_210 |
0.0924206733 |
- |
- |
PREDICTED: THO complex subunit 2 [Jatropha curcas] |
| 12 |
Hb_001473_090 |
0.0927158671 |
- |
- |
PREDICTED: uncharacterized protein LOC105650374 [Jatropha curcas] |
| 13 |
Hb_000347_570 |
0.0930223929 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12b isoform X1 [Jatropha curcas] |
| 14 |
Hb_015099_010 |
0.0930827486 |
- |
- |
PREDICTED: probable glucan 1,3-alpha-glucosidase [Jatropha curcas] |
| 15 |
Hb_102269_010 |
0.0944705207 |
- |
- |
hypothetical protein POPTR_0006s28940g [Populus trichocarpa] |
| 16 |
Hb_001493_040 |
0.0951209698 |
- |
- |
PREDICTED: FACT complex subunit SPT16-like [Jatropha curcas] |
| 17 |
Hb_000363_380 |
0.0953221904 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105633025 [Jatropha curcas] |
| 18 |
Hb_000120_620 |
0.0956227384 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1a isoform X1 [Jatropha curcas] |
| 19 |
Hb_002235_160 |
0.0957510586 |
- |
- |
PREDICTED: SNF1-related protein kinase catalytic subunit alpha KIN10 [Jatropha curcas] |
| 20 |
Hb_010315_090 |
0.0968733276 |
- |
- |
suppressor of ty, putative [Ricinus communis] |