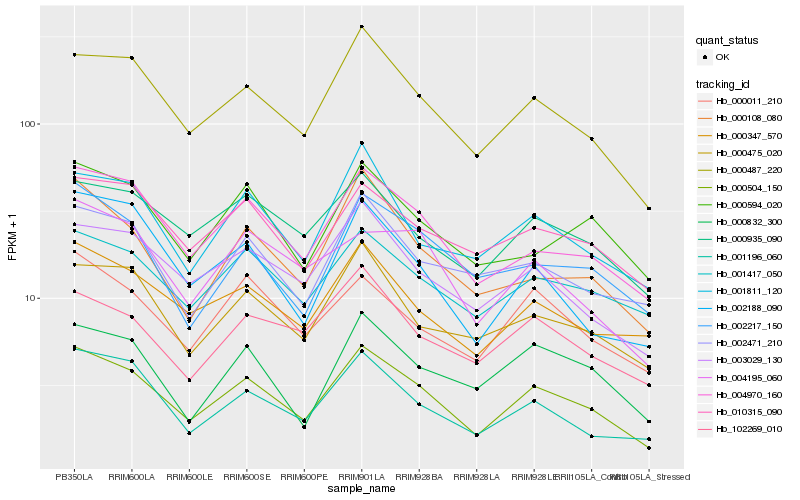
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000504_150 |
0.0 |
- |
- |
PREDICTED: nuclear pore complex protein GP210 [Jatropha curcas] |
| 2 |
Hb_000487_220 |
0.0689334452 |
- |
- |
eukaryotic translation elongation factor, putative [Ricinus communis] |
| 3 |
Hb_004970_160 |
0.0792170981 |
- |
- |
PREDICTED: THO complex subunit 5B-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_003029_130 |
0.0944280374 |
- |
- |
carboxypeptidase regulatory region-containingprotein, putative [Ricinus communis] |
| 5 |
Hb_001196_060 |
0.0945314413 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 6 |
Hb_001811_120 |
0.0996068346 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000347_570 |
0.1003710409 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12b isoform X1 [Jatropha curcas] |
| 8 |
Hb_000475_020 |
0.1006954407 |
- |
- |
PREDICTED: dehydrogenase/reductase SDR family member on chromosome X homolog isoform X2 [Jatropha curcas] |
| 9 |
Hb_000108_080 |
0.1013196939 |
- |
- |
PREDICTED: importin-5 [Jatropha curcas] |
| 10 |
Hb_000011_210 |
0.1022609968 |
- |
- |
PREDICTED: FACT complex subunit SPT16-like [Jatropha curcas] |
| 11 |
Hb_001417_050 |
0.104277125 |
- |
- |
PREDICTED: G patch domain-containing protein TGH [Jatropha curcas] |
| 12 |
Hb_010315_090 |
0.1042779265 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 13 |
Hb_000935_090 |
0.1044156571 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 14 |
Hb_002188_090 |
0.1044228157 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40A isoform X1 [Jatropha curcas] |
| 15 |
Hb_002217_150 |
0.1058617492 |
- |
- |
PREDICTED: nuclear export mediator factor Nemf [Jatropha curcas] |
| 16 |
Hb_000832_300 |
0.1070660204 |
- |
- |
- |
| 17 |
Hb_002471_210 |
0.1088573478 |
- |
- |
PREDICTED: THO complex subunit 2 [Jatropha curcas] |
| 18 |
Hb_000594_020 |
0.1093265443 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 25 isoform X2 [Jatropha curcas] |
| 19 |
Hb_102269_010 |
0.1100522691 |
- |
- |
hypothetical protein POPTR_0006s28940g [Populus trichocarpa] |
| 20 |
Hb_004195_060 |
0.1104190802 |
- |
- |
PREDICTED: uncharacterized protein LOC105803780 isoform X1 [Gossypium raimondii] |