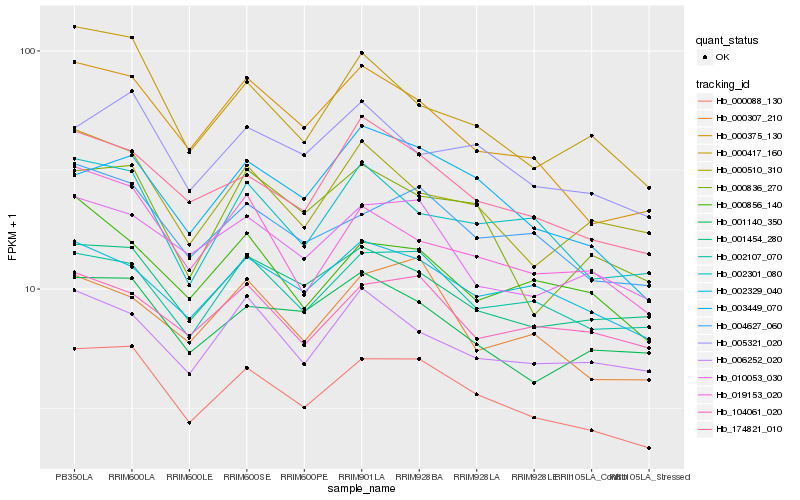
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000088_130 |
0.0 |
- |
- |
hypothetical protein JCGZ_02447 [Jatropha curcas] |
| 2 |
Hb_004627_060 |
0.0646342295 |
- |
- |
PREDICTED: ethylene receptor isoform X2 [Jatropha curcas] |
| 3 |
Hb_000375_130 |
0.0707549342 |
- |
- |
Leucine-rich repeat transmembrane protein kinase family protein isoform 1 [Theobroma cacao] |
| 4 |
Hb_174821_010 |
0.072555379 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 14 [Jatropha curcas] |
| 5 |
Hb_002107_070 |
0.0726419962 |
- |
- |
hypothetical protein RCOM_1598630 [Ricinus communis] |
| 6 |
Hb_001454_280 |
0.0788320814 |
- |
- |
PREDICTED: chaperone protein dnaJ 49 [Jatropha curcas] |
| 7 |
Hb_002301_080 |
0.0801149018 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105649589 isoform X2 [Jatropha curcas] |
| 8 |
Hb_010053_030 |
0.0803855567 |
- |
- |
PREDICTED: tRNA (cytosine(34)-C(5))-methyltransferase [Jatropha curcas] |
| 9 |
Hb_003449_070 |
0.081147607 |
transcription factor |
TF Family: SNF2 |
DNA repair helicase rad5,16, putative [Ricinus communis] |
| 10 |
Hb_005321_020 |
0.0825786903 |
- |
- |
splicing factor-related family protein [Populus trichocarpa] |
| 11 |
Hb_002329_040 |
0.0826307079 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 12 |
Hb_000307_210 |
0.0834588273 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 13 |
Hb_000417_160 |
0.0858296841 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g45780 [Jatropha curcas] |
| 14 |
Hb_000836_270 |
0.0866503804 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog MMK2 [Jatropha curcas] |
| 15 |
Hb_019153_020 |
0.0867005527 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 16 |
Hb_001140_350 |
0.0882147508 |
- |
- |
PREDICTED: uncharacterized protein LOC105648465 [Jatropha curcas] |
| 17 |
Hb_104061_020 |
0.0891893195 |
- |
- |
PREDICTED: UV-stimulated scaffold protein A homolog [Jatropha curcas] |
| 18 |
Hb_000856_140 |
0.0894199103 |
- |
- |
PREDICTED: uncharacterized protein LOC105640478 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000510_310 |
0.090427993 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like [Jatropha curcas] |
| 20 |
Hb_006252_020 |
0.0915024741 |
- |
- |
PREDICTED: SET and MYND domain-containing protein 4 isoform X2 [Jatropha curcas] |