| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
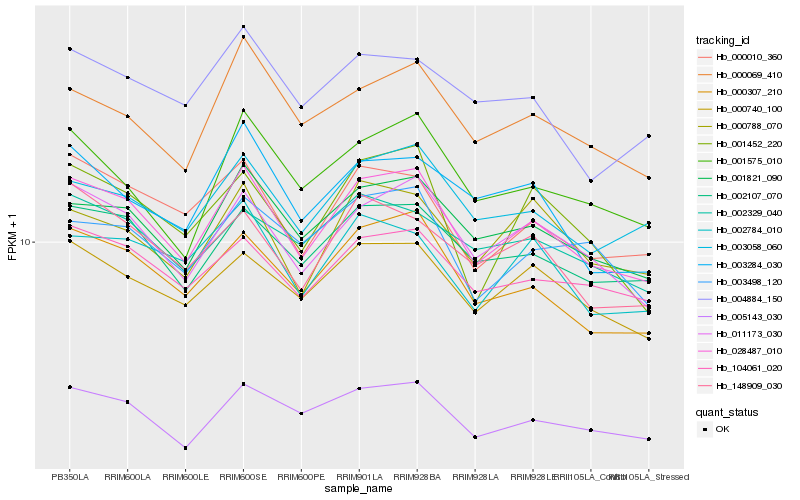
Hb_028487_010 |
0.0 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 23 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000307_210 |
0.0504435908 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 3 |
Hb_011173_030 |
0.0508438999 |
- |
- |
pumilio, putative [Ricinus communis] |
| 4 |
Hb_001821_090 |
0.0576164613 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 5 |
Hb_002107_070 |
0.0586070393 |
- |
- |
hypothetical protein RCOM_1598630 [Ricinus communis] |
| 6 |
Hb_000740_100 |
0.0636833683 |
- |
- |
calpain, putative [Ricinus communis] |
| 7 |
Hb_005143_030 |
0.0644020571 |
- |
- |
PREDICTED: uncharacterized protein LOC105638843 [Jatropha curcas] |
| 8 |
Hb_000010_360 |
0.0648456493 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY [Jatropha curcas] |
| 9 |
Hb_104061_020 |
0.0691016321 |
- |
- |
PREDICTED: UV-stimulated scaffold protein A homolog [Jatropha curcas] |
| 10 |
Hb_003058_060 |
0.0715071711 |
- |
- |
PREDICTED: proteasome-associated protein ECM29 homolog [Jatropha curcas] |
| 11 |
Hb_000069_410 |
0.0740626245 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 12 |
Hb_148909_030 |
0.0754365483 |
- |
- |
PREDICTED: uncharacterized protein LOC105638192 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001575_010 |
0.0759106111 |
- |
- |
PREDICTED: pumilio homolog 6, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_002784_010 |
0.077182406 |
- |
- |
DNA-directed RNA polymerase I subunit, putative [Ricinus communis] |
| 15 |
Hb_000788_070 |
0.0786744892 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g10270 [Jatropha curcas] |
| 16 |
Hb_001452_220 |
0.08040431 |
- |
- |
PREDICTED: molybdopterin biosynthesis protein CNX1 [Jatropha curcas] |
| 17 |
Hb_003498_120 |
0.0807135046 |
- |
- |
hypothetical protein POPTR_0015s12430g, partial [Populus trichocarpa] |
| 18 |
Hb_003284_030 |
0.0811706739 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002329_040 |
0.081288441 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 20 |
Hb_004884_150 |
0.0813584068 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1-like [Jatropha curcas] |