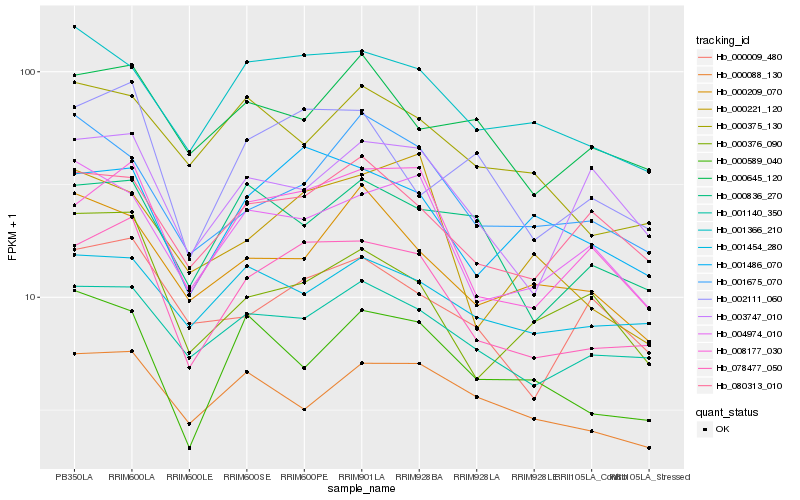
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_078477_050 |
0.0 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105648332 isoform X1 [Jatropha curcas] |
| 2 |
Hb_008177_030 |
0.083333583 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_001486_070 |
0.1077498072 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 7 [Jatropha curcas] |
| 4 |
Hb_001140_350 |
0.1112735724 |
- |
- |
PREDICTED: uncharacterized protein LOC105648465 [Jatropha curcas] |
| 5 |
Hb_001366_210 |
0.1156210444 |
- |
- |
cathepsin B, putative [Ricinus communis] |
| 6 |
Hb_002111_060 |
0.1219664327 |
- |
- |
PREDICTED: toll-like receptor 13 [Jatropha curcas] |
| 7 |
Hb_080313_010 |
0.1228664682 |
- |
- |
PREDICTED: uncharacterized protein LOC105634976 [Jatropha curcas] |
| 8 |
Hb_004974_010 |
0.123246164 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880-like [Prunus mume] |
| 9 |
Hb_000009_480 |
0.1275747227 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g66560 [Jatropha curcas] |
| 10 |
Hb_001454_280 |
0.1316497405 |
- |
- |
PREDICTED: chaperone protein dnaJ 49 [Jatropha curcas] |
| 11 |
Hb_000836_270 |
0.1330522919 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog MMK2 [Jatropha curcas] |
| 12 |
Hb_003747_010 |
0.1348384845 |
- |
- |
hypothetical protein JCGZ_24080 [Jatropha curcas] |
| 13 |
Hb_000376_090 |
0.1348801404 |
- |
- |
Myosin heavy chain, fast skeletal muscle, embryonic, putative [Ricinus communis] |
| 14 |
Hb_000209_070 |
0.1379847163 |
- |
- |
PREDICTED: chloride channel protein CLC-f isoform X1 [Jatropha curcas] |
| 15 |
Hb_000221_120 |
0.1380247506 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 16 |
Hb_001675_070 |
0.138086771 |
- |
- |
eukaryotic translation elongation factor, putative [Ricinus communis] |
| 17 |
Hb_000088_130 |
0.1383350014 |
- |
- |
hypothetical protein JCGZ_02447 [Jatropha curcas] |
| 18 |
Hb_000589_040 |
0.1389035657 |
desease resistance |
Gene Name: zf-BED |
PREDICTED: probable disease resistance protein At1g12280 [Jatropha curcas] |
| 19 |
Hb_000375_130 |
0.1393084355 |
- |
- |
Leucine-rich repeat transmembrane protein kinase family protein isoform 1 [Theobroma cacao] |
| 20 |
Hb_000645_120 |
0.1397217446 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 32 [Jatropha curcas] |