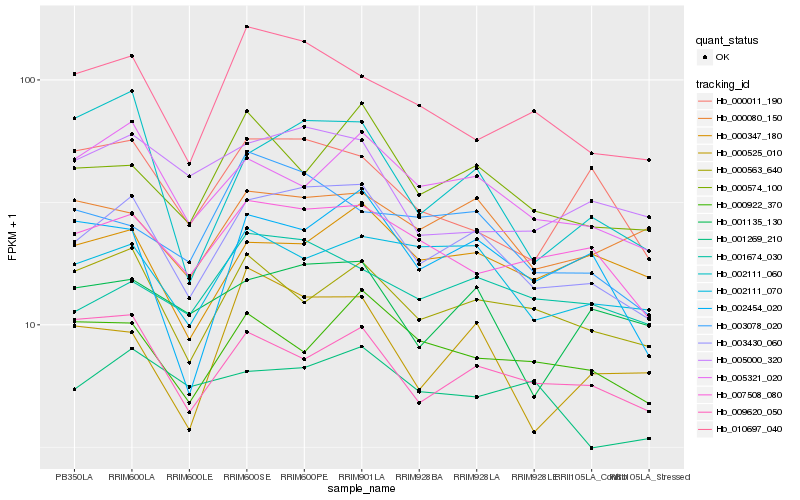
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003430_060 |
0.0 |
- |
- |
PREDICTED: potassium transporter 23 isoform X2 [Elaeis guineensis] |
| 2 |
Hb_007508_080 |
0.0962683877 |
- |
- |
PREDICTED: presenilin-like protein At2g29900 [Jatropha curcas] |
| 3 |
Hb_005321_020 |
0.0983730063 |
- |
- |
splicing factor-related family protein [Populus trichocarpa] |
| 4 |
Hb_000574_100 |
0.1021180315 |
- |
- |
DEAD-box protein abstrakt [Populus trichocarpa] |
| 5 |
Hb_002111_070 |
0.1030470915 |
- |
- |
PREDICTED: protein S-acyltransferase 24 [Jatropha curcas] |
| 6 |
Hb_010697_040 |
0.1064148618 |
- |
- |
PREDICTED: protein GPR107 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000563_640 |
0.1068122741 |
- |
- |
PREDICTED: chaperone protein dnaJ 49 [Jatropha curcas] |
| 8 |
Hb_001674_030 |
0.1087963725 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g24240 [Jatropha curcas] |
| 9 |
Hb_001135_130 |
0.1110689252 |
- |
- |
hypothetical protein CISIN_1g0171752mg, partial [Citrus sinensis] |
| 10 |
Hb_009620_050 |
0.1115181632 |
- |
- |
PREDICTED: protein unc-45 homolog A [Jatropha curcas] |
| 11 |
Hb_000922_370 |
0.1123234435 |
- |
- |
PREDICTED: uncharacterized protein LOC105640366 [Jatropha curcas] |
| 12 |
Hb_003078_020 |
0.1129651251 |
- |
- |
PREDICTED: U-box domain-containing protein 9 [Jatropha curcas] |
| 13 |
Hb_002454_020 |
0.1136529165 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 9 [Vitis vinifera] |
| 14 |
Hb_000347_180 |
0.1150484076 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Jatropha curcas] |
| 15 |
Hb_000080_150 |
0.115311118 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X2 [Jatropha curcas] |
| 16 |
Hb_002111_060 |
0.1158079141 |
- |
- |
PREDICTED: toll-like receptor 13 [Jatropha curcas] |
| 17 |
Hb_001269_210 |
0.115870816 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g76280 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000525_010 |
0.1164647545 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 19 |
Hb_005000_320 |
0.1170120082 |
- |
- |
PREDICTED: casein kinase I isoform delta-like [Jatropha curcas] |
| 20 |
Hb_000011_190 |
0.1177852809 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 6 [Jatropha curcas] |