| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
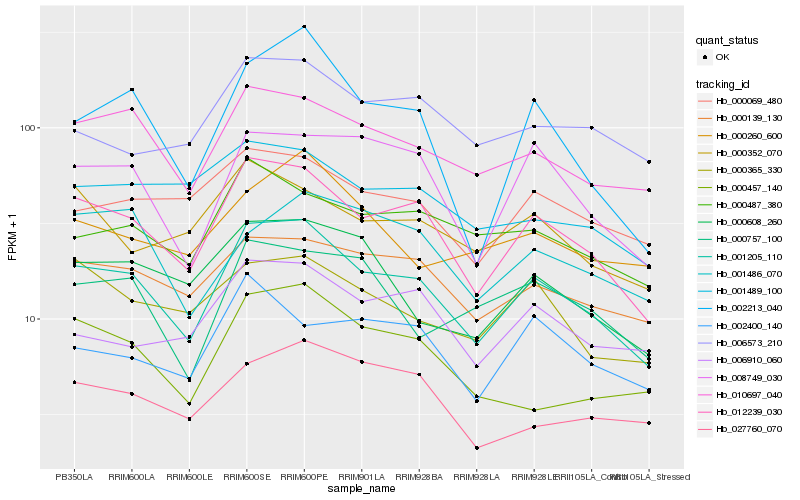
Hb_001205_110 |
0.0 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 4 [Jatropha curcas] |
| 2 |
Hb_012239_030 |
0.0457155113 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_010697_040 |
0.0911727049 |
- |
- |
PREDICTED: protein GPR107 isoform X1 [Jatropha curcas] |
| 4 |
Hb_008749_030 |
0.1045204778 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 5 |
Hb_000139_130 |
0.1048749295 |
- |
- |
PREDICTED: chloride channel protein CLC-d isoform X1 [Jatropha curcas] |
| 6 |
Hb_000608_260 |
0.1121104358 |
- |
- |
PREDICTED: probable protein phosphatase 2C 33 [Jatropha curcas] |
| 7 |
Hb_002213_040 |
0.1135023079 |
- |
- |
cinnamoyl-CoA reductase [Hevea brasiliensis] |
| 8 |
Hb_000487_380 |
0.1171093257 |
- |
- |
PREDICTED: protein PAT1 homolog 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000069_480 |
0.1183029589 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000365_330 |
0.1189193341 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_000457_140 |
0.1194978102 |
- |
- |
PREDICTED: transmembrane protein 136-like [Jatropha curcas] |
| 12 |
Hb_006910_060 |
0.1210354308 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780-like [Jatropha curcas] |
| 13 |
Hb_000757_100 |
0.1221017143 |
- |
- |
SPla/RYanodine receptor domain-containing protein isoform 2 [Theobroma cacao] |
| 14 |
Hb_027760_070 |
0.122574821 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02-like [Jatropha curcas] |
| 15 |
Hb_006573_210 |
0.1231487067 |
- |
- |
PREDICTED: protein SGT1 homolog [Jatropha curcas] |
| 16 |
Hb_001489_100 |
0.1233195044 |
- |
- |
PREDICTED: hypersensitive-induced response protein 2 [Jatropha curcas] |
| 17 |
Hb_000352_070 |
0.1249521053 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 18 |
Hb_001486_070 |
0.1255703622 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 7 [Jatropha curcas] |
| 19 |
Hb_000260_600 |
0.1267696064 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002400_140 |
0.129773729 |
- |
- |
protein kinase, putative [Ricinus communis] |