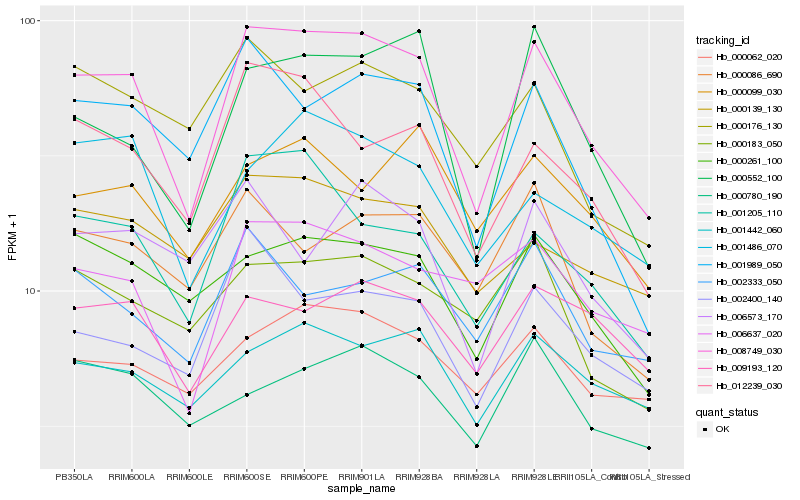
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008749_030 |
0.0 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 2 |
Hb_001205_110 |
0.1045204778 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 4 [Jatropha curcas] |
| 3 |
Hb_012239_030 |
0.1064443081 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_000552_100 |
0.1074877394 |
- |
- |
PREDICTED: nitrile-specifier protein 5-like [Jatropha curcas] |
| 5 |
Hb_001989_050 |
0.1109511038 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 5 [Jatropha curcas] |
| 6 |
Hb_000261_100 |
0.1131667725 |
- |
- |
PREDICTED: beta-galactosidase 9 isoform X2 [Jatropha curcas] |
| 7 |
Hb_002400_140 |
0.1163838645 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 8 |
Hb_006637_020 |
0.1187229919 |
- |
- |
PREDICTED: uncharacterized protein LOC105642234 [Jatropha curcas] |
| 9 |
Hb_001486_070 |
0.118853388 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 7 [Jatropha curcas] |
| 10 |
Hb_001442_060 |
0.1219473554 |
- |
- |
PREDICTED: uncharacterized protein LOC105638091 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000062_020 |
0.122678566 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 12 |
Hb_000139_130 |
0.1256149151 |
- |
- |
PREDICTED: chloride channel protein CLC-d isoform X1 [Jatropha curcas] |
| 13 |
Hb_009193_120 |
0.1257370902 |
desease resistance |
Gene Name: AAA_22 |
PREDICTED: putative ATP-dependent RNA helicase DHX33 [Jatropha curcas] |
| 14 |
Hb_000183_050 |
0.125975396 |
- |
- |
PREDICTED: alpha-mannosidase 2 [Jatropha curcas] |
| 15 |
Hb_002333_050 |
0.1260875306 |
- |
- |
PREDICTED: uncharacterized protein LOC105644854 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000086_690 |
0.1266019365 |
- |
- |
PREDICTED: splicing factor 3B subunit 1 [Jatropha curcas] |
| 17 |
Hb_000780_190 |
0.1276461449 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 2 [Jatropha curcas] |
| 18 |
Hb_006573_170 |
0.128665697 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000176_130 |
0.128992839 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Jatropha curcas] |
| 20 |
Hb_000099_030 |
0.129140291 |
- |
- |
6-phosphogluconate dehydrogenase, putative [Ricinus communis] |