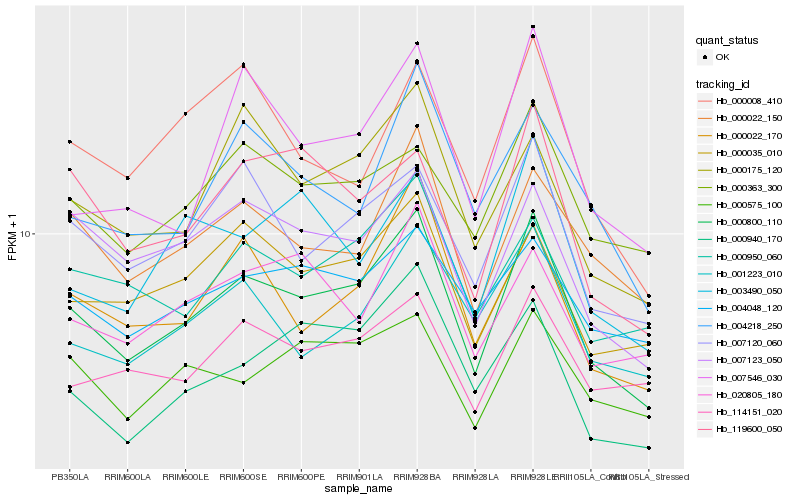
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000800_110 |
0.0 |
- |
- |
NAD dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_007120_060 |
0.1068541484 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 3 |
Hb_000035_010 |
0.1109046337 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR12-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000022_170 |
0.1115409547 |
- |
- |
PREDICTED: uncharacterized protein LOC105628473 [Jatropha curcas] |
| 5 |
Hb_000022_150 |
0.1117175618 |
- |
- |
- |
| 6 |
Hb_007123_050 |
0.1130514405 |
- |
- |
Vitellogenic carboxypeptidase, putative [Ricinus communis] |
| 7 |
Hb_000950_060 |
0.1168174102 |
- |
- |
PREDICTED: CCAAT/enhancer-binding protein zeta [Jatropha curcas] |
| 8 |
Hb_001223_010 |
0.1172235605 |
- |
- |
PREDICTED: FGGY carbohydrate kinase domain-containing protein isoform X2 [Jatropha curcas] |
| 9 |
Hb_007546_030 |
0.1175168762 |
- |
- |
transcription cofactor, putative [Ricinus communis] |
| 10 |
Hb_000175_120 |
0.1181770842 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 11 |
Hb_114151_020 |
0.1186288018 |
- |
- |
glucan water dikinase 2 [Manihot esculenta] |
| 12 |
Hb_000575_100 |
0.1208747211 |
- |
- |
PREDICTED: nephrocystin-3 isoform X2 [Jatropha curcas] |
| 13 |
Hb_004218_250 |
0.122181475 |
- |
- |
PREDICTED: beta-galactosidase [Jatropha curcas] |
| 14 |
Hb_000363_300 |
0.1222315581 |
- |
- |
PREDICTED: protein ENHANCED DOWNY MILDEW 2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_119600_050 |
0.1227160573 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 16 |
Hb_020805_180 |
0.1274895088 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 17 |
Hb_003490_050 |
0.1279062126 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 20, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000940_170 |
0.1296666633 |
- |
- |
hypothetical protein VITISV_029834 [Vitis vinifera] |
| 19 |
Hb_000008_410 |
0.130220075 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase [Hevea brasiliensis] |
| 20 |
Hb_004048_120 |
0.1303383735 |
- |
- |
PREDICTED: nuclear pore complex protein NUP160 isoform X3 [Jatropha curcas] |