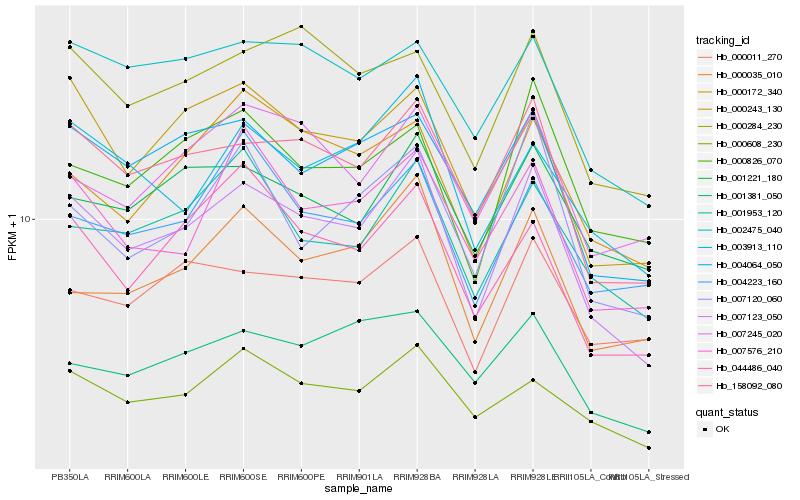
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007123_050 |
0.0 |
- |
- |
Vitellogenic carboxypeptidase, putative [Ricinus communis] |
| 2 |
Hb_158092_080 |
0.0643715367 |
- |
- |
hypothetical protein JCGZ_23092 [Jatropha curcas] |
| 3 |
Hb_004064_050 |
0.085116391 |
- |
- |
Structural maintenance of chromosome, putative [Ricinus communis] |
| 4 |
Hb_000243_130 |
0.0919575534 |
- |
- |
hypothetical protein M569_10795, partial [Genlisea aurea] |
| 5 |
Hb_000608_230 |
0.093797717 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 6 |
Hb_044486_040 |
0.0958156401 |
- |
- |
PREDICTED: ion channel CASTOR-like [Malus domestica] |
| 7 |
Hb_003913_110 |
0.0962028134 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |
| 8 |
Hb_000035_010 |
0.0964551508 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR12-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_002475_040 |
0.0966084658 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 10 |
Hb_007576_210 |
0.0978805052 |
- |
- |
PREDICTED: protein furry homolog [Jatropha curcas] |
| 11 |
Hb_000284_230 |
0.0983622199 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |
| 12 |
Hb_001953_120 |
0.0990635086 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001221_180 |
0.1022595684 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 14 |
Hb_001381_050 |
0.1025054631 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 15 |
Hb_000826_070 |
0.1032652456 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000172_340 |
0.1032805832 |
- |
- |
PREDICTED: uncharacterized protein LOC101261327 [Solanum lycopersicum] |
| 17 |
Hb_000011_270 |
0.1044380988 |
- |
- |
PREDICTED: pumilio homolog 23 [Jatropha curcas] |
| 18 |
Hb_007120_060 |
0.1046579169 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 19 |
Hb_004223_160 |
0.1049720731 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X2 [Jatropha curcas] |
| 20 |
Hb_007245_020 |
0.1050224341 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |