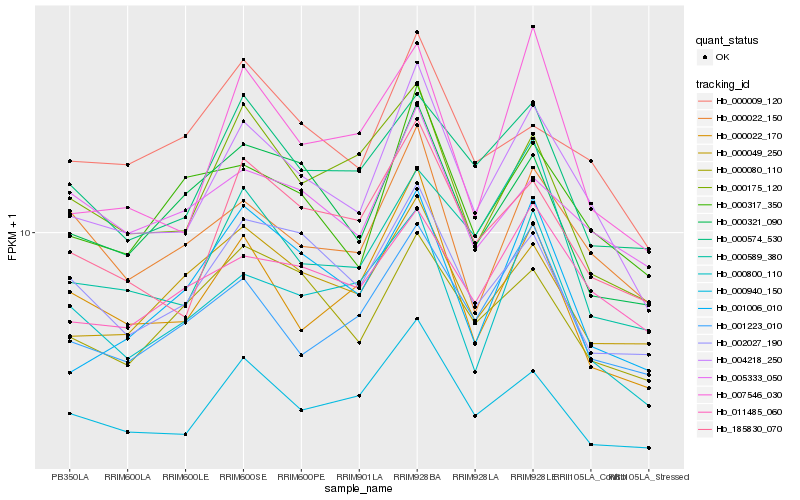
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004218_250 |
0.0 |
- |
- |
PREDICTED: beta-galactosidase [Jatropha curcas] |
| 2 |
Hb_185830_070 |
0.093555734 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000940_150 |
0.0979995258 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |
| 4 |
Hb_000317_350 |
0.1076508561 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3 [Jatropha curcas] |
| 5 |
Hb_000022_170 |
0.1104440656 |
- |
- |
PREDICTED: uncharacterized protein LOC105628473 [Jatropha curcas] |
| 6 |
Hb_000022_150 |
0.1122553657 |
- |
- |
- |
| 7 |
Hb_002027_190 |
0.1150900992 |
- |
- |
PREDICTED: autophagy-related protein 18g [Jatropha curcas] |
| 8 |
Hb_001223_010 |
0.1154270936 |
- |
- |
PREDICTED: FGGY carbohydrate kinase domain-containing protein isoform X2 [Jatropha curcas] |
| 9 |
Hb_000321_090 |
0.1179457946 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 10 |
Hb_000049_250 |
0.1182529457 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 11 |
Hb_011485_060 |
0.118309065 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 3 [Jatropha curcas] |
| 12 |
Hb_000009_120 |
0.1187298849 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P isoform X1 [Jatropha curcas] |
| 13 |
Hb_000574_530 |
0.1187679523 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 14 |
Hb_000175_120 |
0.1191570039 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 15 |
Hb_007546_030 |
0.120414391 |
- |
- |
transcription cofactor, putative [Ricinus communis] |
| 16 |
Hb_001006_010 |
0.121524691 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 17 |
Hb_000800_110 |
0.122181475 |
- |
- |
NAD dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_000080_110 |
0.122406937 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000589_380 |
0.1232263025 |
- |
- |
Phosphoribosylformylglycinamidine synthase, putative [Ricinus communis] |
| 20 |
Hb_005333_050 |
0.126166246 |
- |
- |
PREDICTED: glucose-6-phosphate isomerase 1, chloroplastic [Jatropha curcas] |