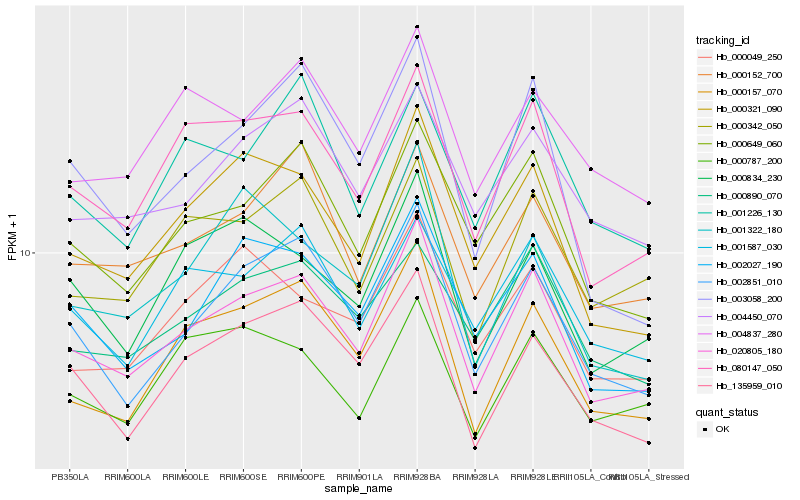
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020805_180 |
0.0 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 2 |
Hb_080147_050 |
0.0651657818 |
- |
- |
PREDICTED: probable plastidic glucose transporter 2 [Jatropha curcas] |
| 3 |
Hb_000321_090 |
0.0666043953 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 4 |
Hb_002851_010 |
0.0691762769 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 5 |
Hb_000649_060 |
0.069733858 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIE1 [Jatropha curcas] |
| 6 |
Hb_000157_070 |
0.0734811128 |
- |
- |
PREDICTED: alpha-N-acetylglucosaminidase [Jatropha curcas] |
| 7 |
Hb_002027_190 |
0.076745613 |
- |
- |
PREDICTED: autophagy-related protein 18g [Jatropha curcas] |
| 8 |
Hb_000152_700 |
0.081120196 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIN2 [Jatropha curcas] |
| 9 |
Hb_135959_010 |
0.0815455818 |
- |
- |
hypothetical protein JCGZ_07060 [Jatropha curcas] |
| 10 |
Hb_000342_050 |
0.0823323646 |
- |
- |
hypothetical protein POPTR_0006s24710g [Populus trichocarpa] |
| 11 |
Hb_004450_070 |
0.0826879923 |
- |
- |
PREDICTED: 2-hydroxyacyl-CoA lyase [Jatropha curcas] |
| 12 |
Hb_001587_030 |
0.0864641582 |
- |
- |
hypothetical protein JCGZ_09892 [Jatropha curcas] |
| 13 |
Hb_000890_070 |
0.0885751221 |
- |
- |
PREDICTED: protein ALWAYS EARLY 3 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000834_230 |
0.088684652 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 15 |
Hb_004837_280 |
0.0909442868 |
- |
- |
PREDICTED: uncharacterized protein LOC105648296 [Jatropha curcas] |
| 16 |
Hb_000049_250 |
0.0912668607 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003058_200 |
0.0935488932 |
- |
- |
PREDICTED: dihydropyrimidinase [Populus euphratica] |
| 18 |
Hb_001226_130 |
0.0964742175 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 19 |
Hb_000787_200 |
0.0966964973 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_001322_180 |
0.0971473014 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |