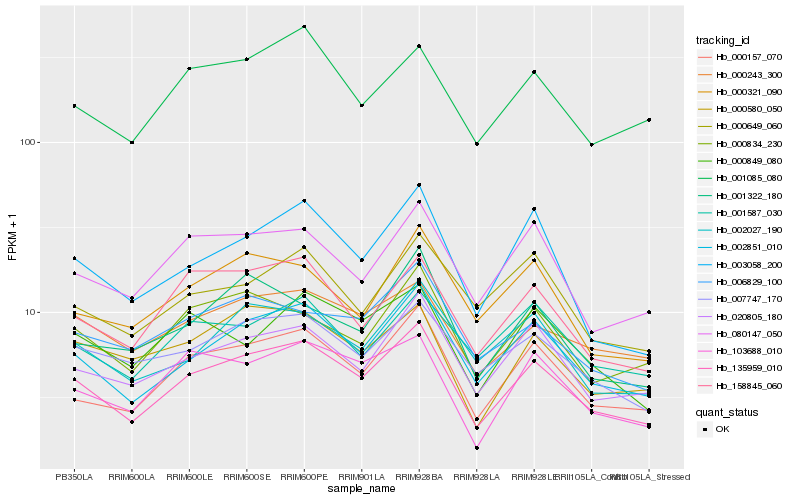
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_135959_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_07060 [Jatropha curcas] |
| 2 |
Hb_002851_010 |
0.0547490636 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 3 |
Hb_000157_070 |
0.0702356906 |
- |
- |
PREDICTED: alpha-N-acetylglucosaminidase [Jatropha curcas] |
| 4 |
Hb_020805_180 |
0.0815455818 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 5 |
Hb_158845_060 |
0.0909928783 |
- |
- |
PREDICTED: nudix hydrolase 3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002027_190 |
0.0929019455 |
- |
- |
PREDICTED: autophagy-related protein 18g [Jatropha curcas] |
| 7 |
Hb_007747_170 |
0.0932377189 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 8 |
Hb_000834_230 |
0.0934453599 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 9 |
Hb_003058_200 |
0.0940292267 |
- |
- |
PREDICTED: dihydropyrimidinase [Populus euphratica] |
| 10 |
Hb_080147_050 |
0.0941681788 |
- |
- |
PREDICTED: probable plastidic glucose transporter 2 [Jatropha curcas] |
| 11 |
Hb_000243_300 |
0.0952081323 |
- |
- |
catalytic, putative [Ricinus communis] |
| 12 |
Hb_103688_010 |
0.0956001105 |
- |
- |
PREDICTED: sucrose-phosphatase 2-like [Jatropha curcas] |
| 13 |
Hb_006829_100 |
0.098281648 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000580_050 |
0.1004656894 |
- |
- |
- |
| 15 |
Hb_000321_090 |
0.1006008762 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 16 |
Hb_001085_080 |
0.1014177668 |
- |
- |
eukaryotic translation elongation factor 1-alpha [Hevea brasiliensis] |
| 17 |
Hb_001587_030 |
0.1045646075 |
- |
- |
hypothetical protein JCGZ_09892 [Jatropha curcas] |
| 18 |
Hb_001322_180 |
0.1057378289 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |
| 19 |
Hb_000649_060 |
0.1059007763 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIE1 [Jatropha curcas] |
| 20 |
Hb_000849_080 |
0.1064014494 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKU5 isoform X1 [Jatropha curcas] |