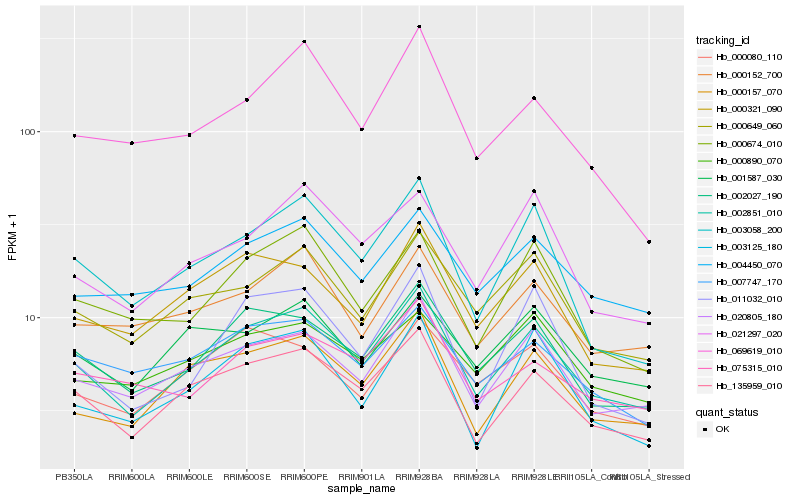
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002851_010 |
0.0 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 2 |
Hb_135959_010 |
0.0547490636 |
- |
- |
hypothetical protein JCGZ_07060 [Jatropha curcas] |
| 3 |
Hb_002027_190 |
0.0667953381 |
- |
- |
PREDICTED: autophagy-related protein 18g [Jatropha curcas] |
| 4 |
Hb_020805_180 |
0.0691762769 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 5 |
Hb_000157_070 |
0.0787564747 |
- |
- |
PREDICTED: alpha-N-acetylglucosaminidase [Jatropha curcas] |
| 6 |
Hb_000649_060 |
0.0802296672 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIE1 [Jatropha curcas] |
| 7 |
Hb_003058_200 |
0.0862550544 |
- |
- |
PREDICTED: dihydropyrimidinase [Populus euphratica] |
| 8 |
Hb_004450_070 |
0.0871147464 |
- |
- |
PREDICTED: 2-hydroxyacyl-CoA lyase [Jatropha curcas] |
| 9 |
Hb_000674_010 |
0.0880427527 |
- |
- |
PREDICTED: copper-transporting ATPase RAN1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000321_090 |
0.0910128841 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 11 |
Hb_021297_020 |
0.09296557 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 12 |
Hb_000152_700 |
0.0954583003 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIN2 [Jatropha curcas] |
| 13 |
Hb_075315_010 |
0.0972507602 |
- |
- |
ornithine aminotransferase [Camellia sinensis] |
| 14 |
Hb_007747_170 |
0.0973002492 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 15 |
Hb_003125_180 |
0.0976732714 |
- |
- |
hypothetical protein JCGZ_26440 [Jatropha curcas] |
| 16 |
Hb_001587_030 |
0.0987775051 |
- |
- |
hypothetical protein JCGZ_09892 [Jatropha curcas] |
| 17 |
Hb_011032_010 |
0.0994290219 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 18 |
Hb_000080_110 |
0.1006529345 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_069619_010 |
0.1008856082 |
- |
- |
Aspartic proteinase precursor, putative [Ricinus communis] |
| 20 |
Hb_000890_070 |
0.104038659 |
- |
- |
PREDICTED: protein ALWAYS EARLY 3 isoform X2 [Jatropha curcas] |