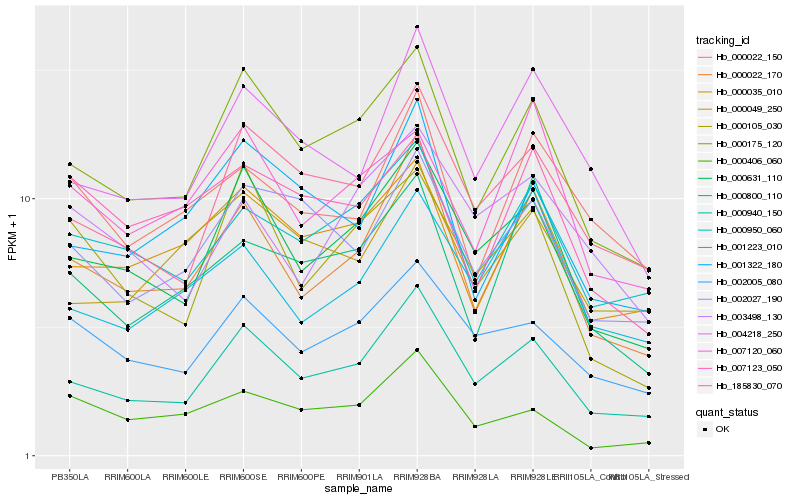
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000022_170 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628473 [Jatropha curcas] |
| 2 |
Hb_000940_150 |
0.077095198 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |
| 3 |
Hb_000175_120 |
0.0957160668 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 4 |
Hb_000631_110 |
0.0986690136 |
- |
- |
PREDICTED: activating signal cointegrator 1 complex subunit 3 [Jatropha curcas] |
| 5 |
Hb_000950_060 |
0.1094023488 |
- |
- |
PREDICTED: CCAAT/enhancer-binding protein zeta [Jatropha curcas] |
| 6 |
Hb_004218_250 |
0.1104440656 |
- |
- |
PREDICTED: beta-galactosidase [Jatropha curcas] |
| 7 |
Hb_000800_110 |
0.1115409547 |
- |
- |
NAD dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_000022_150 |
0.1135441292 |
- |
- |
- |
| 9 |
Hb_001223_010 |
0.1176679398 |
- |
- |
PREDICTED: FGGY carbohydrate kinase domain-containing protein isoform X2 [Jatropha curcas] |
| 10 |
Hb_000035_010 |
0.1177626915 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR12-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_001322_180 |
0.1247764422 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 26 [Jatropha curcas] |
| 12 |
Hb_185830_070 |
0.1292970151 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_007120_060 |
0.1311412562 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 14 |
Hb_002005_080 |
0.1347541883 |
- |
- |
PREDICTED: uncharacterized protein LOC105649750 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000105_030 |
0.1393048482 |
- |
- |
fkbp-rapamycin associated protein, putative [Ricinus communis] |
| 16 |
Hb_002027_190 |
0.140318469 |
- |
- |
PREDICTED: autophagy-related protein 18g [Jatropha curcas] |
| 17 |
Hb_000406_060 |
0.1407062335 |
- |
- |
hypothetical protein CICLE_v10000115mg [Citrus clementina] |
| 18 |
Hb_003498_130 |
0.1409777569 |
- |
- |
PREDICTED: protein ROOT HAIR DEFECTIVE 3 homolog 2-like [Citrus sinensis] |
| 19 |
Hb_007123_050 |
0.1416253967 |
- |
- |
Vitellogenic carboxypeptidase, putative [Ricinus communis] |
| 20 |
Hb_000049_250 |
0.1418322434 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |