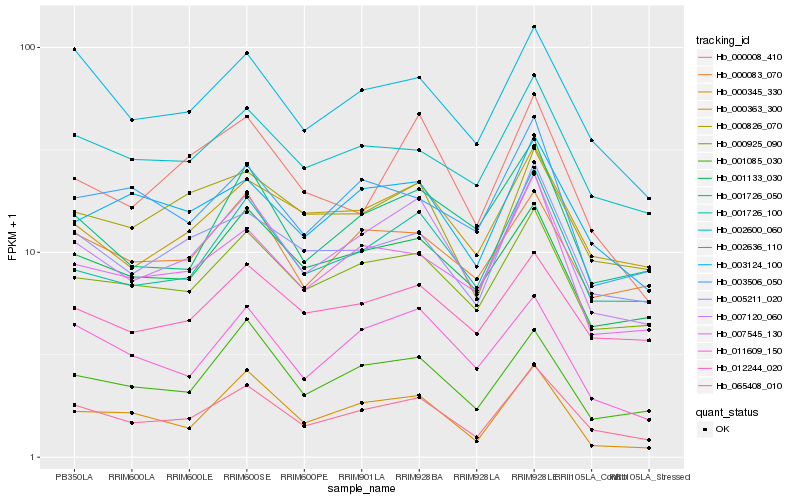
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_065408_010 |
0.0 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 2 |
Hb_007120_060 |
0.0634215934 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 3 |
Hb_005211_020 |
0.0814684379 |
- |
- |
PREDICTED: serine/threonine-protein kinase prpf4B-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_002636_110 |
0.0826250501 |
- |
- |
PREDICTED: heat shock protein 83-like isoform X2 [Gossypium raimondii] |
| 5 |
Hb_001726_100 |
0.0836538086 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 19 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000925_090 |
0.0844562397 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001085_030 |
0.0911807441 |
- |
- |
Tetratricopeptide repeat-like superfamily protein, putative isoform 1 [Theobroma cacao] |
| 8 |
Hb_007545_130 |
0.0914789583 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002600_060 |
0.0928876754 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 10 |
Hb_001133_030 |
0.0983965529 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 11 |
Hb_000826_070 |
0.1012667968 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003124_100 |
0.1021623498 |
- |
- |
- |
| 13 |
Hb_000083_070 |
0.1034552191 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000363_300 |
0.1047177596 |
- |
- |
PREDICTED: protein ENHANCED DOWNY MILDEW 2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_011609_150 |
0.104820747 |
- |
- |
PREDICTED: HEAT repeat-containing protein 5B [Jatropha curcas] |
| 16 |
Hb_001726_050 |
0.1058765313 |
transcription factor |
TF Family: C3H |
Nucleic acid binding,zinc ion binding,DNA binding, putative isoform 1 [Theobroma cacao] |
| 17 |
Hb_012244_020 |
0.1070948108 |
- |
- |
calpain, putative [Ricinus communis] |
| 18 |
Hb_000008_410 |
0.1111622065 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase [Hevea brasiliensis] |
| 19 |
Hb_003506_050 |
0.1123011852 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_000345_330 |
0.1135500002 |
desease resistance |
Gene Name: NB-ARC |
NB-ARC domain-containing disease resistance-like protein isoform 1 [Theobroma cacao] |