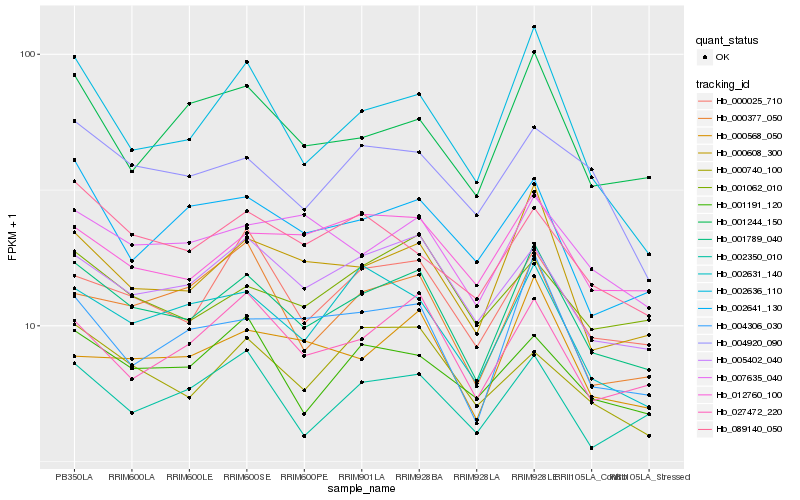
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001789_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648457 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000608_300 |
0.063691817 |
- |
- |
PREDICTED: protein KRI1 homolog [Jatropha curcas] |
| 3 |
Hb_005402_040 |
0.0677046925 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |
| 4 |
Hb_004306_030 |
0.0724213599 |
- |
- |
PREDICTED: GPI transamidase component PIG-S [Jatropha curcas] |
| 5 |
Hb_027472_220 |
0.0725249803 |
- |
- |
PREDICTED: nucleolar complex protein 3 homolog isoform X1 [Jatropha curcas] |
| 6 |
Hb_000025_710 |
0.0726805638 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 7 |
Hb_002631_140 |
0.0762914206 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 12, chloroplastic [Jatropha curcas] |
| 8 |
Hb_002350_010 |
0.078676628 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit rpc1 [Jatropha curcas] |
| 9 |
Hb_007635_040 |
0.0798563235 |
- |
- |
PREDICTED: WAT1-related protein At3g45870 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001244_150 |
0.079929594 |
- |
- |
transformer serine/arginine-rich ribonucleoprotein [Populus trichocarpa] |
| 11 |
Hb_012760_100 |
0.0802175404 |
- |
- |
Pre-rRNA-processing protein ESF1, putative [Ricinus communis] |
| 12 |
Hb_000568_050 |
0.081111197 |
- |
- |
PREDICTED: endoplasmic reticulum metallopeptidase 1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001191_120 |
0.0822704815 |
- |
- |
PREDICTED: cleavage stimulating factor 64 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002636_110 |
0.0838626215 |
- |
- |
PREDICTED: heat shock protein 83-like isoform X2 [Gossypium raimondii] |
| 15 |
Hb_000740_100 |
0.0839776239 |
- |
- |
calpain, putative [Ricinus communis] |
| 16 |
Hb_089140_050 |
0.0841117757 |
- |
- |
hypothetical protein JCGZ_07485 [Jatropha curcas] |
| 17 |
Hb_001062_010 |
0.0845415816 |
- |
- |
PREDICTED: CWF19-like protein 2 [Jatropha curcas] |
| 18 |
Hb_000377_050 |
0.0846235419 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 19 |
Hb_004920_090 |
0.0853104065 |
- |
- |
PREDICTED: probable glutamyl endopeptidase, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_002641_130 |
0.0853710012 |
- |
- |
PREDICTED: uncharacterized protein LOC105643556 [Jatropha curcas] |