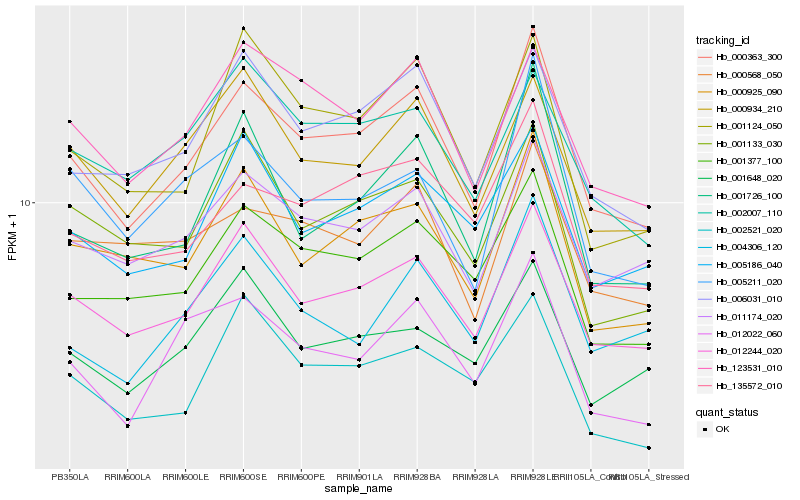
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000363_300 |
0.0 |
- |
- |
PREDICTED: protein ENHANCED DOWNY MILDEW 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_012244_020 |
0.0499603018 |
- |
- |
calpain, putative [Ricinus communis] |
| 3 |
Hb_011174_020 |
0.0626564465 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 4 |
Hb_001377_100 |
0.0648685277 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 5 |
Hb_006031_010 |
0.0650218462 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15b isoform X3 [Jatropha curcas] |
| 6 |
Hb_135572_010 |
0.0680930234 |
- |
- |
GTP-binding protein alpha subunit, gna, putative [Ricinus communis] |
| 7 |
Hb_012022_060 |
0.0682103583 |
- |
- |
PREDICTED: uncharacterized protein LOC105646588 isoform X2 [Jatropha curcas] |
| 8 |
Hb_123531_010 |
0.0695031628 |
- |
- |
PREDICTED: nuclear pore complex protein NUP98A isoform X1 [Jatropha curcas] |
| 9 |
Hb_000925_090 |
0.0707816127 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001726_100 |
0.0713894855 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 19 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000934_210 |
0.0727850745 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 12 |
Hb_005186_040 |
0.0734096434 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 13 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001124_050 |
0.0789884037 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromatin structure-remodeling complex protein SYD-like [Jatropha curcas] |
| 14 |
Hb_004306_120 |
0.0791196835 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_001133_030 |
0.0793025678 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 16 |
Hb_001648_020 |
0.0794614727 |
- |
- |
hypothetical protein POPTR_0007s07750g [Populus trichocarpa] |
| 17 |
Hb_000568_050 |
0.0803541446 |
- |
- |
PREDICTED: endoplasmic reticulum metallopeptidase 1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_005211_020 |
0.0808613052 |
- |
- |
PREDICTED: serine/threonine-protein kinase prpf4B-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_002521_020 |
0.0808702571 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g03100, mitochondrial-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_002007_110 |
0.0826877846 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |