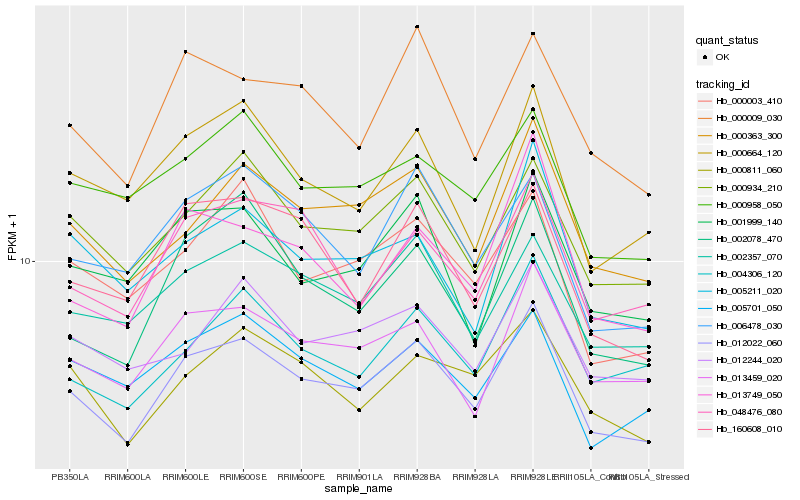
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012022_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646588 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000363_300 |
0.0682103583 |
- |
- |
PREDICTED: protein ENHANCED DOWNY MILDEW 2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000934_210 |
0.0747879508 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 4 |
Hb_013459_020 |
0.0774245391 |
- |
- |
PREDICTED: uncharacterized protein LOC105628886 [Jatropha curcas] |
| 5 |
Hb_002357_070 |
0.0796402536 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 6 |
Hb_000009_030 |
0.0830295266 |
- |
- |
PREDICTED: splicing factor U2af large subunit B isoform X1 [Jatropha curcas] |
| 7 |
Hb_000664_120 |
0.0841611497 |
- |
- |
PREDICTED: SART-1 family protein DOT2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000811_060 |
0.0850059665 |
- |
- |
PREDICTED: uncharacterized protein LOC105650696 [Jatropha curcas] |
| 9 |
Hb_160608_010 |
0.0867071187 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 10 |
Hb_001999_140 |
0.0873427496 |
- |
- |
PREDICTED: anthranilate synthase alpha subunit 2, chloroplastic isoform X2 [Jatropha curcas] |
| 11 |
Hb_002078_470 |
0.0875083304 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 4 isoform X1 [Jatropha curcas] |
| 12 |
Hb_004306_120 |
0.0875288299 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_006478_030 |
0.0881814444 |
- |
- |
hypothetical protein POPTR_0005s23510g [Populus trichocarpa] |
| 14 |
Hb_048476_080 |
0.0882555017 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 15 |
Hb_000958_050 |
0.0889133209 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 16 |
Hb_000003_410 |
0.0889189487 |
- |
- |
PREDICTED: intron-binding protein aquarius [Jatropha curcas] |
| 17 |
Hb_012244_020 |
0.0900608292 |
- |
- |
calpain, putative [Ricinus communis] |
| 18 |
Hb_013749_050 |
0.0907960054 |
- |
- |
PREDICTED: uncharacterized protein LOC105647765 isoform X2 [Jatropha curcas] |
| 19 |
Hb_005211_020 |
0.0912001777 |
- |
- |
PREDICTED: serine/threonine-protein kinase prpf4B-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_005701_050 |
0.0914115499 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |