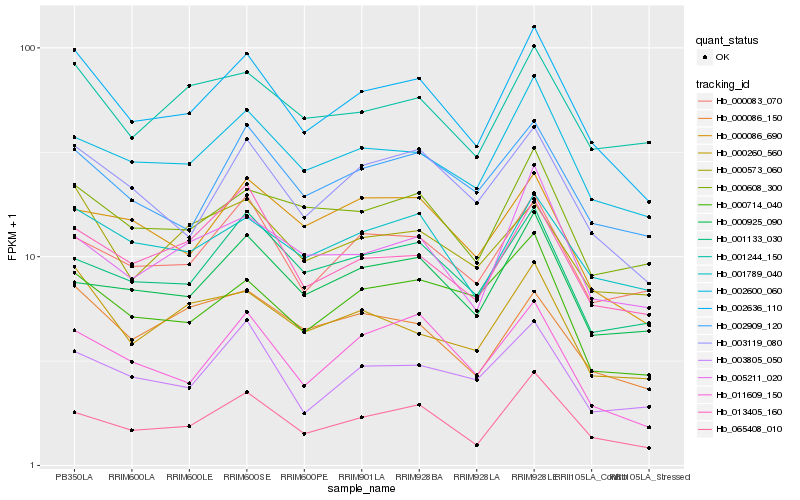
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002636_110 |
0.0 |
- |
- |
PREDICTED: heat shock protein 83-like isoform X2 [Gossypium raimondii] |
| 2 |
Hb_003119_080 |
0.076704054 |
- |
- |
PREDICTED: splicing factor 3B subunit 1 [Jatropha curcas] |
| 3 |
Hb_002600_060 |
0.0771329692 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 4 |
Hb_065408_010 |
0.0826250501 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 5 |
Hb_001789_040 |
0.0838626215 |
- |
- |
PREDICTED: uncharacterized protein LOC105648457 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002909_120 |
0.0875011853 |
- |
- |
PREDICTED: transcription elongation factor SPT6 [Jatropha curcas] |
| 7 |
Hb_000608_300 |
0.0881842944 |
- |
- |
PREDICTED: protein KRI1 homolog [Jatropha curcas] |
| 8 |
Hb_000260_560 |
0.0916049007 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g18110, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000083_070 |
0.0921140527 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000573_060 |
0.0935731256 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2D isoform X1 [Jatropha curcas] |
| 11 |
Hb_000714_040 |
0.0938443034 |
- |
- |
PREDICTED: uncharacterized protein LOC105634664 isoform X2 [Jatropha curcas] |
| 12 |
Hb_011609_150 |
0.0947379073 |
- |
- |
PREDICTED: HEAT repeat-containing protein 5B [Jatropha curcas] |
| 13 |
Hb_001244_150 |
0.0950530544 |
- |
- |
transformer serine/arginine-rich ribonucleoprotein [Populus trichocarpa] |
| 14 |
Hb_001133_030 |
0.0955266722 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 15 |
Hb_005211_020 |
0.0958004061 |
- |
- |
PREDICTED: serine/threonine-protein kinase prpf4B-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000086_150 |
0.0970633331 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X1 [Jatropha curcas] |
| 17 |
Hb_013405_160 |
0.0970634243 |
- |
- |
PREDICTED: methyltransferase-like protein 1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000925_090 |
0.0977540761 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003805_050 |
0.0996551235 |
- |
- |
- |
| 20 |
Hb_000086_690 |
0.0997864191 |
- |
- |
PREDICTED: splicing factor 3B subunit 1 [Jatropha curcas] |