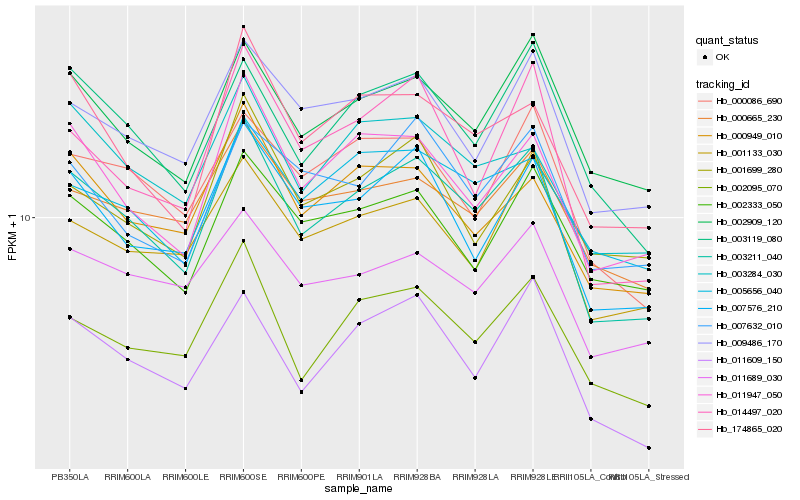
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003211_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634664 isoform X1 [Jatropha curcas] |
| 2 |
Hb_007576_210 |
0.0717687631 |
- |
- |
PREDICTED: protein furry homolog [Jatropha curcas] |
| 3 |
Hb_002095_070 |
0.0727350223 |
- |
- |
PREDICTED: probable pre-mRNA-splicing factor ATP-dependent RNA helicase [Jatropha curcas] |
| 4 |
Hb_011947_050 |
0.0728653831 |
- |
- |
E3 ubiquitin protein ligase upl2, putative [Ricinus communis] |
| 5 |
Hb_003119_080 |
0.0749145045 |
- |
- |
PREDICTED: splicing factor 3B subunit 1 [Jatropha curcas] |
| 6 |
Hb_002909_120 |
0.0752380644 |
- |
- |
PREDICTED: transcription elongation factor SPT6 [Jatropha curcas] |
| 7 |
Hb_009486_170 |
0.0753671984 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 8 |
Hb_174865_020 |
0.076593191 |
- |
- |
E3 ubiquitin protein ligase upl2, putative [Ricinus communis] |
| 9 |
Hb_014497_020 |
0.0778107441 |
- |
- |
PREDICTED: U5 small nuclear ribonucleoprotein 200 kDa helicase [Jatropha curcas] |
| 10 |
Hb_011609_150 |
0.0796816 |
- |
- |
PREDICTED: HEAT repeat-containing protein 5B [Jatropha curcas] |
| 11 |
Hb_003284_030 |
0.0821700644 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000949_010 |
0.0826119082 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF2 [Jatropha curcas] |
| 13 |
Hb_011689_030 |
0.0833389765 |
- |
- |
80 kD MCM3-associated protein, putative [Ricinus communis] |
| 14 |
Hb_000086_690 |
0.0841512101 |
- |
- |
PREDICTED: splicing factor 3B subunit 1 [Jatropha curcas] |
| 15 |
Hb_001133_030 |
0.0847864416 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 16 |
Hb_000665_230 |
0.0865833168 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 17 |
Hb_005656_040 |
0.0867659125 |
desease resistance |
Gene Name: T2SE |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 18 |
Hb_002333_050 |
0.0870751422 |
- |
- |
PREDICTED: uncharacterized protein LOC105644854 isoform X1 [Jatropha curcas] |
| 19 |
Hb_007632_010 |
0.089436554 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001699_280 |
0.0943085909 |
- |
- |
PREDICTED: transformation/transcription domain-associated protein [Jatropha curcas] |