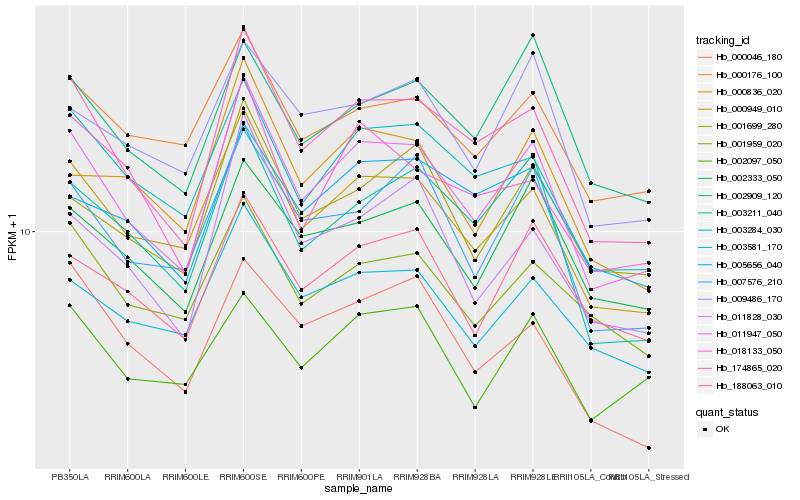
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011947_050 |
0.0 |
- |
- |
E3 ubiquitin protein ligase upl2, putative [Ricinus communis] |
| 2 |
Hb_174865_020 |
0.0392970863 |
- |
- |
E3 ubiquitin protein ligase upl2, putative [Ricinus communis] |
| 3 |
Hb_000949_010 |
0.0519327199 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF2 [Jatropha curcas] |
| 4 |
Hb_003284_030 |
0.0713144364 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_003211_040 |
0.0728653831 |
- |
- |
PREDICTED: uncharacterized protein LOC105634664 isoform X1 [Jatropha curcas] |
| 6 |
Hb_007576_210 |
0.0739618726 |
- |
- |
PREDICTED: protein furry homolog [Jatropha curcas] |
| 7 |
Hb_002333_050 |
0.0757006593 |
- |
- |
PREDICTED: uncharacterized protein LOC105644854 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001699_280 |
0.0759300625 |
- |
- |
PREDICTED: transformation/transcription domain-associated protein [Jatropha curcas] |
| 9 |
Hb_003581_170 |
0.0764374578 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 10 |
Hb_011828_030 |
0.0779870107 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 11 |
Hb_188063_010 |
0.0784445594 |
- |
- |
hypothetical protein RCOM_1469330 [Ricinus communis] |
| 12 |
Hb_000836_020 |
0.0813120812 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 13 |
Hb_000176_100 |
0.0822422064 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Solanum lycopersicum] |
| 14 |
Hb_001959_020 |
0.0823459964 |
- |
- |
PREDICTED: uncharacterized protein LOC105638454 isoform X1 [Jatropha curcas] |
| 15 |
Hb_009486_170 |
0.0851145691 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 16 |
Hb_002909_120 |
0.0866645869 |
- |
- |
PREDICTED: transcription elongation factor SPT6 [Jatropha curcas] |
| 17 |
Hb_018133_050 |
0.0871684588 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 1 [Jatropha curcas] |
| 18 |
Hb_005656_040 |
0.0877966489 |
desease resistance |
Gene Name: T2SE |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 19 |
Hb_002097_050 |
0.088125933 |
- |
- |
PREDICTED: protein fluG isoform X1 [Jatropha curcas] |
| 20 |
Hb_000046_180 |
0.0902637253 |
- |
- |
PREDICTED: uncharacterized protein LOC105631840 [Jatropha curcas] |