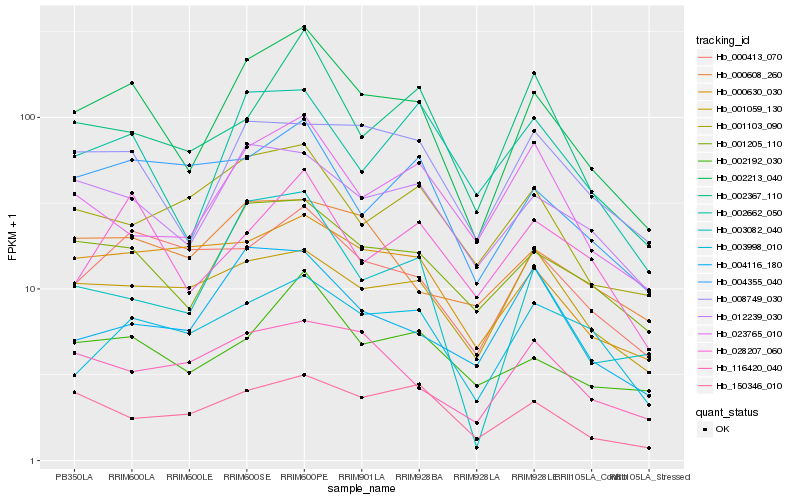
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002213_040 |
0.0 |
- |
- |
cinnamoyl-CoA reductase [Hevea brasiliensis] |
| 2 |
Hb_001205_110 |
0.1135023079 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 4 [Jatropha curcas] |
| 3 |
Hb_012239_030 |
0.1333376047 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_002192_030 |
0.1401924273 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_008749_030 |
0.1465640663 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 6 |
Hb_003082_040 |
0.1469858949 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Phoenix dactylifera] |
| 7 |
Hb_002662_050 |
0.1498620478 |
- |
- |
cysteine protease CP1 [Manihot esculenta] |
| 8 |
Hb_000413_070 |
0.1516776538 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_028207_060 |
0.1569674998 |
- |
- |
PREDICTED: uncharacterized protein LOC105639235 [Jatropha curcas] |
| 10 |
Hb_004116_180 |
0.1590547313 |
- |
- |
PREDICTED: uncharacterized protein LOC105646147 [Jatropha curcas] |
| 11 |
Hb_002367_110 |
0.1594636939 |
- |
- |
PREDICTED: cinnamyl alcohol dehydrogenase 1 [Sesamum indicum] |
| 12 |
Hb_000630_030 |
0.1599271847 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2 isoform X1 [Populus euphratica] |
| 13 |
Hb_023765_010 |
0.1599732048 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 14 |
Hb_003998_010 |
0.1608299731 |
- |
- |
PREDICTED: protein BRASSINOSTEROID INSENSITIVE 1 [Jatropha curcas] |
| 15 |
Hb_000608_260 |
0.1633177326 |
- |
- |
PREDICTED: probable protein phosphatase 2C 33 [Jatropha curcas] |
| 16 |
Hb_001103_090 |
0.1640463718 |
- |
- |
PREDICTED: patellin-3 [Populus euphratica] |
| 17 |
Hb_004355_040 |
0.1644883758 |
- |
- |
epoxide hydrolase, putative [Ricinus communis] |
| 18 |
Hb_001059_130 |
0.164588273 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_150346_010 |
0.1646011216 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 20 |
Hb_116420_040 |
0.1652370215 |
- |
- |
conserved hypothetical protein [Ricinus communis] |