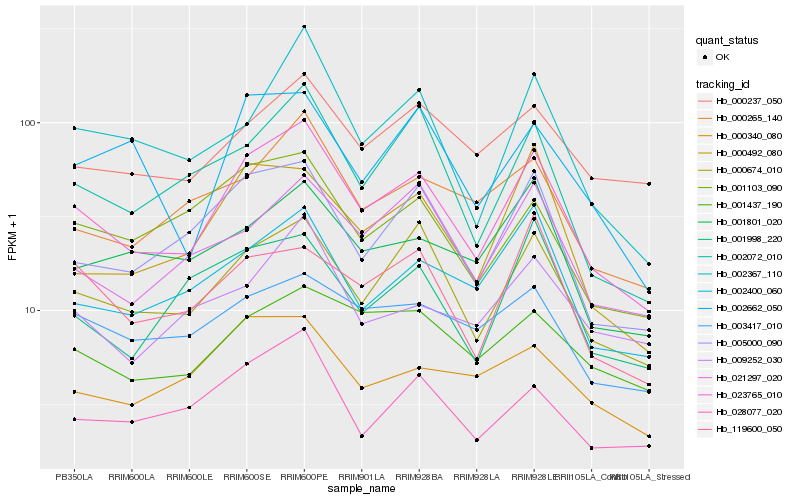
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_023765_010 |
0.0 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 2 |
Hb_000674_010 |
0.0902954543 |
- |
- |
PREDICTED: copper-transporting ATPase RAN1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_005000_090 |
0.0998378117 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 4 |
Hb_002072_010 |
0.1115808109 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002400_060 |
0.1160684908 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 6 |
Hb_028077_020 |
0.1160793125 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002367_110 |
0.1161832624 |
- |
- |
PREDICTED: cinnamyl alcohol dehydrogenase 1 [Sesamum indicum] |
| 8 |
Hb_000265_140 |
0.1179466762 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 12 [Jatropha curcas] |
| 9 |
Hb_000492_080 |
0.1181623991 |
- |
- |
PREDICTED: probable sucrose-phosphate synthase 1 [Jatropha curcas] |
| 10 |
Hb_001103_090 |
0.1203537604 |
- |
- |
PREDICTED: patellin-3 [Populus euphratica] |
| 11 |
Hb_021297_020 |
0.1208326411 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 12 |
Hb_001437_190 |
0.1273183881 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 13 |
Hb_000237_050 |
0.1275910339 |
- |
- |
CP2 [Hevea brasiliensis] |
| 14 |
Hb_001801_020 |
0.1291151143 |
- |
- |
PREDICTED: xylosyltransferase 1 [Jatropha curcas] |
| 15 |
Hb_001998_220 |
0.129892593 |
- |
- |
PREDICTED: isoamylase 1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_119600_050 |
0.1300570927 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 17 |
Hb_003417_010 |
0.1306989578 |
- |
- |
PREDICTED: protein TPLATE [Jatropha curcas] |
| 18 |
Hb_000340_080 |
0.1309863955 |
- |
- |
PREDICTED: isoamylase 2, chloroplastic [Jatropha curcas] |
| 19 |
Hb_009252_030 |
0.1332009203 |
- |
- |
PREDICTED: CASP-like protein 4A3 [Jatropha curcas] |
| 20 |
Hb_002662_050 |
0.1341340332 |
- |
- |
cysteine protease CP1 [Manihot esculenta] |