| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
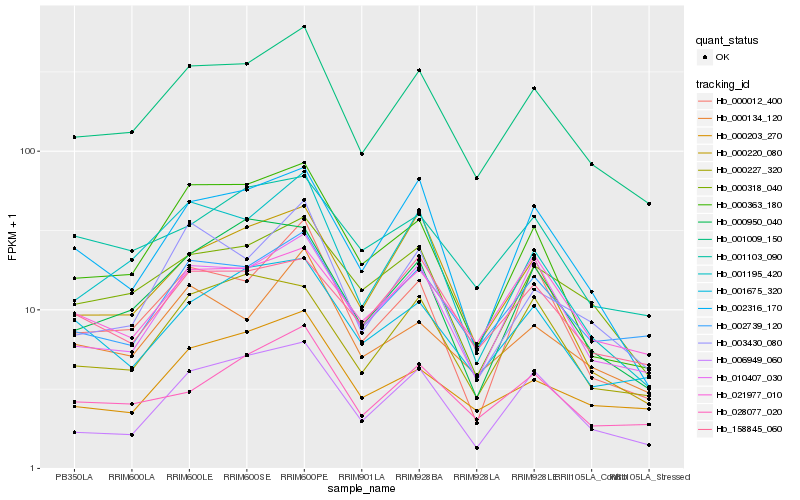
Hb_001009_150 |
0.0 |
- |
- |
Glyceraldehyde-3-phosphate dehydrogenase [Gossypium arboreum] |
| 2 |
Hb_002739_120 |
0.082458819 |
- |
- |
PREDICTED: zinc finger protein ZPR1-like [Jatropha curcas] |
| 3 |
Hb_010407_030 |
0.0911678378 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 4 |
Hb_001195_420 |
0.094419915 |
- |
- |
PREDICTED: uncharacterized protein LOC105633774 isoform X4 [Jatropha curcas] |
| 5 |
Hb_000318_040 |
0.0946913323 |
- |
- |
hypothetical protein JCGZ_04877 [Jatropha curcas] |
| 6 |
Hb_006949_060 |
0.0995443354 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 7 |
Hb_028077_020 |
0.1010162531 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000134_120 |
0.1049762244 |
- |
- |
PREDICTED: uncharacterized protein LOC105628669 [Jatropha curcas] |
| 9 |
Hb_000203_270 |
0.1059132596 |
- |
- |
alpha-xylosidase, putative [Ricinus communis] |
| 10 |
Hb_003430_080 |
0.1060367569 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_001675_320 |
0.1082708265 |
- |
- |
PREDICTED: uncharacterized protein LOC105635025 [Jatropha curcas] |
| 12 |
Hb_000012_400 |
0.1082967233 |
- |
- |
PREDICTED: K(+) efflux antiporter 6 [Jatropha curcas] |
| 13 |
Hb_000363_180 |
0.1084460482 |
- |
- |
PREDICTED: uncharacterized protein LOC105633052 [Jatropha curcas] |
| 14 |
Hb_002316_170 |
0.1085502652 |
- |
- |
purine permease, putative [Ricinus communis] |
| 15 |
Hb_000950_040 |
0.1123273835 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 16 |
Hb_158845_060 |
0.1124977166 |
- |
- |
PREDICTED: nudix hydrolase 3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001103_090 |
0.1146881914 |
- |
- |
PREDICTED: patellin-3 [Populus euphratica] |
| 18 |
Hb_000227_320 |
0.1153126942 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_000220_080 |
0.1158402559 |
- |
- |
kinase, putative [Ricinus communis] |
| 20 |
Hb_021977_010 |
0.1177773958 |
- |
- |
PREDICTED: leukotriene A-4 hydrolase homolog [Jatropha curcas] |