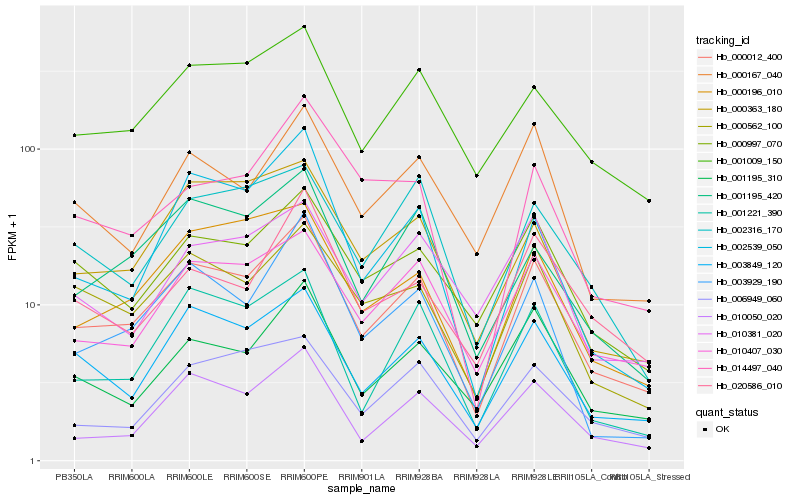
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000012_400 |
0.0 |
- |
- |
PREDICTED: K(+) efflux antiporter 6 [Jatropha curcas] |
| 2 |
Hb_003849_120 |
0.0972487294 |
- |
- |
PREDICTED: uncharacterized protein LOC100264440 [Vitis vinifera] |
| 3 |
Hb_010050_020 |
0.1001850942 |
- |
- |
PREDICTED: uncharacterized protein LOC102601222 [Solanum tuberosum] |
| 4 |
Hb_010407_030 |
0.1019245014 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 5 |
Hb_000562_100 |
0.1054260033 |
- |
- |
PREDICTED: protein IQ-DOMAIN 32 [Jatropha curcas] |
| 6 |
Hb_001195_310 |
0.1082369706 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_001009_150 |
0.1082967233 |
- |
- |
Glyceraldehyde-3-phosphate dehydrogenase [Gossypium arboreum] |
| 8 |
Hb_000997_070 |
0.1117495358 |
- |
- |
PREDICTED: ureidoglycolate hydrolase isoform X1 [Jatropha curcas] |
| 9 |
Hb_000196_010 |
0.1125601035 |
- |
- |
Patellin-5, putative [Ricinus communis] |
| 10 |
Hb_003929_190 |
0.116184396 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g18500 [Jatropha curcas] |
| 11 |
Hb_001195_420 |
0.1161890105 |
- |
- |
PREDICTED: uncharacterized protein LOC105633774 isoform X4 [Jatropha curcas] |
| 12 |
Hb_001221_390 |
0.1171634765 |
- |
- |
Phospholipase C 4 precursor, putative [Ricinus communis] |
| 13 |
Hb_000167_040 |
0.1236879668 |
- |
- |
PREDICTED: serine carboxypeptidase-like [Jatropha curcas] |
| 14 |
Hb_000363_180 |
0.1237565544 |
- |
- |
PREDICTED: uncharacterized protein LOC105633052 [Jatropha curcas] |
| 15 |
Hb_006949_060 |
0.1268259668 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 16 |
Hb_020586_010 |
0.1321239606 |
- |
- |
Alpha-N-arabinofuranosidase 1 precursor, putative [Ricinus communis] |
| 17 |
Hb_002316_170 |
0.1326926497 |
- |
- |
purine permease, putative [Ricinus communis] |
| 18 |
Hb_002539_050 |
0.1332283985 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 19 |
Hb_010381_020 |
0.1343676037 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 20 |
Hb_014497_040 |
0.1345701613 |
- |
- |
PREDICTED: calvin cycle protein CP12-3, chloroplastic [Jatropha curcas] |