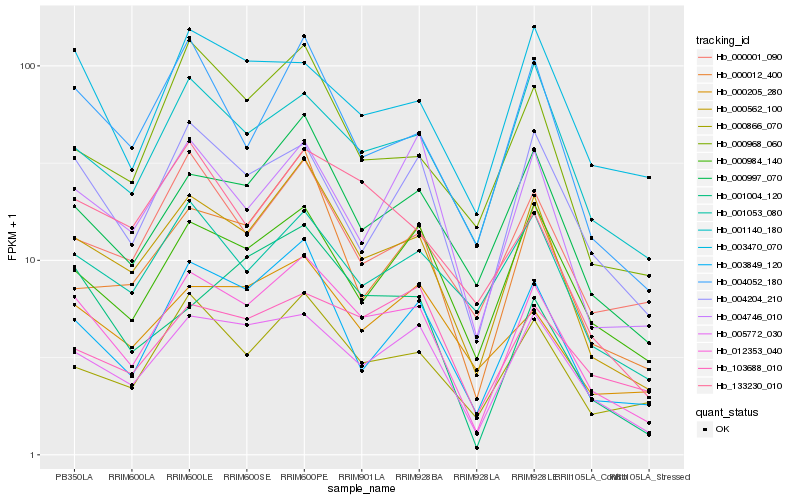
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012353_040 |
0.0 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 2 |
Hb_000562_100 |
0.0788629838 |
- |
- |
PREDICTED: protein IQ-DOMAIN 32 [Jatropha curcas] |
| 3 |
Hb_005772_030 |
0.0982665648 |
- |
- |
PREDICTED: protein trichome birefringence-like 35 [Jatropha curcas] |
| 4 |
Hb_000997_070 |
0.1210377052 |
- |
- |
PREDICTED: ureidoglycolate hydrolase isoform X1 [Jatropha curcas] |
| 5 |
Hb_003849_120 |
0.1220138387 |
- |
- |
PREDICTED: uncharacterized protein LOC100264440 [Vitis vinifera] |
| 6 |
Hb_004204_210 |
0.1244308674 |
- |
- |
PREDICTED: heparanase-like protein 1 [Jatropha curcas] |
| 7 |
Hb_000866_070 |
0.1260706396 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: SKP1-like protein 21 [Jatropha curcas] |
| 8 |
Hb_000984_140 |
0.1307211421 |
- |
- |
PREDICTED: xylulose kinase [Jatropha curcas] |
| 9 |
Hb_004052_180 |
0.1318607526 |
- |
- |
PREDICTED: protein COBRA-like [Jatropha curcas] |
| 10 |
Hb_001140_180 |
0.1350471362 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 11 |
Hb_001053_080 |
0.137900882 |
- |
- |
OsCesA3 protein [Morus notabilis] |
| 12 |
Hb_000205_280 |
0.1394775859 |
- |
- |
PREDICTED: inositol phosphorylceramide glucuronosyltransferase 1 [Jatropha curcas] |
| 13 |
Hb_004746_010 |
0.1438298003 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 14 |
Hb_000001_090 |
0.1462088056 |
- |
- |
PREDICTED: uncharacterized protein At3g58460 [Populus euphratica] |
| 15 |
Hb_003470_070 |
0.147403478 |
- |
- |
rubisco subunit binding-protein beta subunit, rubb, putative [Ricinus communis] |
| 16 |
Hb_000968_060 |
0.1503667696 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |
| 17 |
Hb_103688_010 |
0.1519704323 |
- |
- |
PREDICTED: sucrose-phosphatase 2-like [Jatropha curcas] |
| 18 |
Hb_133230_010 |
0.152424108 |
- |
- |
hypothetical protein L484_001846 [Morus notabilis] |
| 19 |
Hb_000012_400 |
0.1534873405 |
- |
- |
PREDICTED: K(+) efflux antiporter 6 [Jatropha curcas] |
| 20 |
Hb_001004_120 |
0.1535336033 |
- |
- |
PREDICTED: receptor-like protein kinase HERK 1 isoform X2 [Jatropha curcas] |