| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
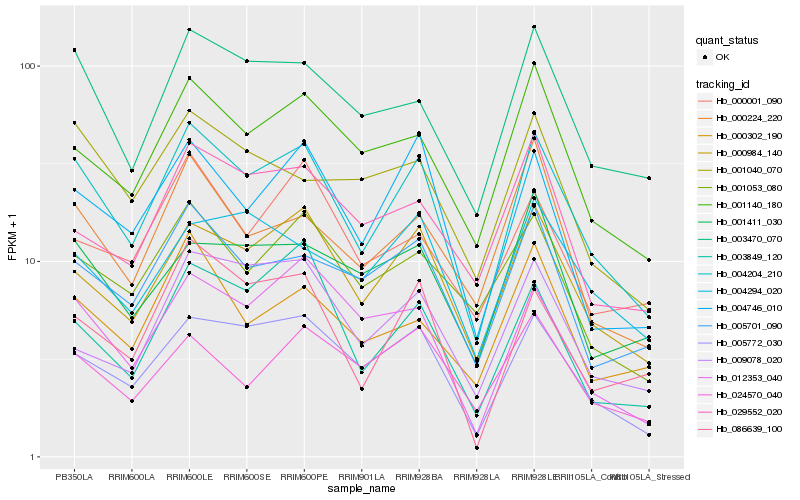
Hb_004204_210 |
0.0 |
- |
- |
PREDICTED: heparanase-like protein 1 [Jatropha curcas] |
| 2 |
Hb_000984_140 |
0.0958228577 |
- |
- |
PREDICTED: xylulose kinase [Jatropha curcas] |
| 3 |
Hb_005772_030 |
0.096608429 |
- |
- |
PREDICTED: protein trichome birefringence-like 35 [Jatropha curcas] |
| 4 |
Hb_003470_070 |
0.1116924808 |
- |
- |
rubisco subunit binding-protein beta subunit, rubb, putative [Ricinus communis] |
| 5 |
Hb_004746_010 |
0.1144271387 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 6 |
Hb_001140_180 |
0.1188229144 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 7 |
Hb_005701_090 |
0.1193896752 |
- |
- |
PREDICTED: protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 6, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_012353_040 |
0.1244308674 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 9 |
Hb_001053_080 |
0.1250352568 |
- |
- |
OsCesA3 protein [Morus notabilis] |
| 10 |
Hb_086639_100 |
0.1268639008 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105646787 [Jatropha curcas] |
| 11 |
Hb_000224_220 |
0.1319495228 |
- |
- |
PREDICTED: protein disulfide isomerase-like 2-3 [Jatropha curcas] |
| 12 |
Hb_003849_120 |
0.1325992735 |
- |
- |
PREDICTED: uncharacterized protein LOC100264440 [Vitis vinifera] |
| 13 |
Hb_000302_190 |
0.137808263 |
- |
- |
PREDICTED: uncharacterized protein At3g49140-like [Jatropha curcas] |
| 14 |
Hb_001040_070 |
0.1390162541 |
- |
- |
PREDICTED: uncharacterized protein LOC105649523 [Jatropha curcas] |
| 15 |
Hb_024570_040 |
0.1399293636 |
- |
- |
PREDICTED: clathrin interactor EPSIN 1 [Jatropha curcas] |
| 16 |
Hb_004294_020 |
0.1414301607 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 17 |
Hb_029552_020 |
0.1418723407 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001411_030 |
0.1435693254 |
- |
- |
hypothetical protein POPTR_0003s09620g [Populus trichocarpa] |
| 19 |
Hb_000001_090 |
0.1446960804 |
- |
- |
PREDICTED: uncharacterized protein At3g58460 [Populus euphratica] |
| 20 |
Hb_009078_020 |
0.1449013506 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |