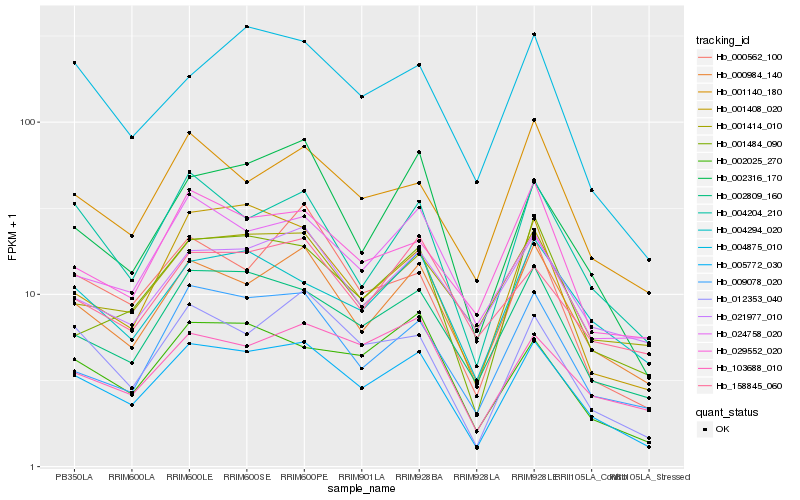
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005772_030 |
0.0 |
- |
- |
PREDICTED: protein trichome birefringence-like 35 [Jatropha curcas] |
| 2 |
Hb_000984_140 |
0.0792495911 |
- |
- |
PREDICTED: xylulose kinase [Jatropha curcas] |
| 3 |
Hb_002809_160 |
0.09205085 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 4 |
Hb_004204_210 |
0.096608429 |
- |
- |
PREDICTED: heparanase-like protein 1 [Jatropha curcas] |
| 5 |
Hb_012353_040 |
0.0982665648 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 6 |
Hb_002025_270 |
0.1088503843 |
- |
- |
PREDICTED: uncharacterized protein At4g15970 isoform X1 [Jatropha curcas] |
| 7 |
Hb_009078_020 |
0.109364045 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 8 |
Hb_001140_180 |
0.1118952676 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 9 |
Hb_004294_020 |
0.1119703093 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 10 |
Hb_103688_010 |
0.113219715 |
- |
- |
PREDICTED: sucrose-phosphatase 2-like [Jatropha curcas] |
| 11 |
Hb_001484_090 |
0.1138559044 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 12 |
Hb_001414_010 |
0.114975358 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 13 |
Hb_002316_170 |
0.1154739544 |
- |
- |
purine permease, putative [Ricinus communis] |
| 14 |
Hb_021977_010 |
0.1161310539 |
- |
- |
PREDICTED: leukotriene A-4 hydrolase homolog [Jatropha curcas] |
| 15 |
Hb_004875_010 |
0.1169854159 |
- |
- |
plasma membrane aquaporin 1 [Hevea brasiliensis] |
| 16 |
Hb_001408_020 |
0.1172855471 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 2-like [Jatropha curcas] |
| 17 |
Hb_024758_020 |
0.1175724646 |
- |
- |
acyl-CoA binding protein 3B [Vernicia fordii] |
| 18 |
Hb_029552_020 |
0.1198738038 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000562_100 |
0.1219644868 |
- |
- |
PREDICTED: protein IQ-DOMAIN 32 [Jatropha curcas] |
| 20 |
Hb_158845_060 |
0.1241655838 |
- |
- |
PREDICTED: nudix hydrolase 3 isoform X1 [Jatropha curcas] |