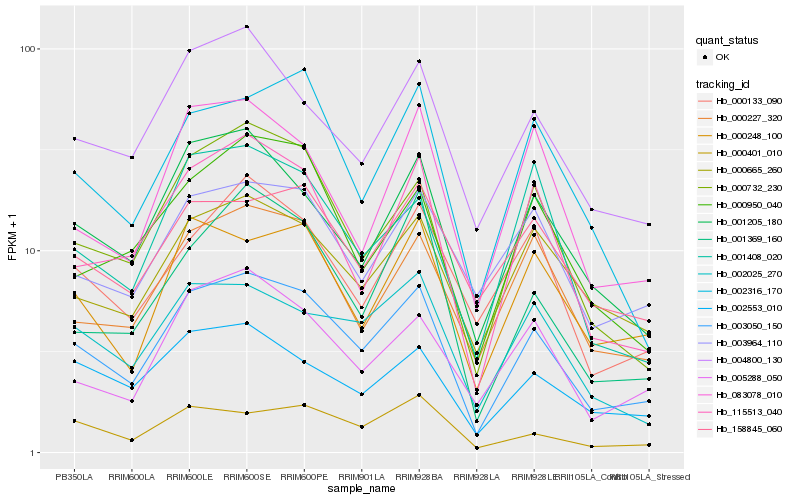
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003050_150 |
0.0 |
- |
- |
endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase, putative [Ricinus communis] |
| 2 |
Hb_000732_230 |
0.1006505794 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_001205_180 |
0.1045957627 |
- |
- |
hypothetical protein POPTR_0005s12580g [Populus trichocarpa] |
| 4 |
Hb_115513_040 |
0.1089929662 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004800_130 |
0.1118573536 |
- |
- |
hypothetical protein POPTR_0005s12580g [Populus trichocarpa] |
| 6 |
Hb_002025_270 |
0.1161836522 |
- |
- |
PREDICTED: uncharacterized protein At4g15970 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002553_010 |
0.1163144466 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 2-like [Populus euphratica] |
| 8 |
Hb_000665_260 |
0.1165268723 |
- |
- |
PREDICTED: uncharacterized protein LOC105637620 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000401_010 |
0.1166108144 |
- |
- |
SAB, putative [Ricinus communis] |
| 10 |
Hb_000248_100 |
0.1173094602 |
- |
- |
PREDICTED: NADPH-dependent thioredoxin reductase 3 [Populus euphratica] |
| 11 |
Hb_000227_320 |
0.1192690052 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_005288_050 |
0.1210193849 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR4 [Jatropha curcas] |
| 13 |
Hb_000950_040 |
0.1248844736 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 14 |
Hb_002316_170 |
0.124976126 |
- |
- |
purine permease, putative [Ricinus communis] |
| 15 |
Hb_000133_090 |
0.1265562409 |
- |
- |
PREDICTED: uncharacterized protein LOC105629320 [Jatropha curcas] |
| 16 |
Hb_003964_110 |
0.1266103311 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g26460, mitochondrial [Jatropha curcas] |
| 17 |
Hb_083078_010 |
0.1276510125 |
- |
- |
Splicing factor, arginine/serine-rich, putative [Ricinus communis] |
| 18 |
Hb_001369_160 |
0.1283761948 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 19 |
Hb_001408_020 |
0.1304337363 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 2-like [Jatropha curcas] |
| 20 |
Hb_158845_060 |
0.1308323634 |
- |
- |
PREDICTED: nudix hydrolase 3 isoform X1 [Jatropha curcas] |