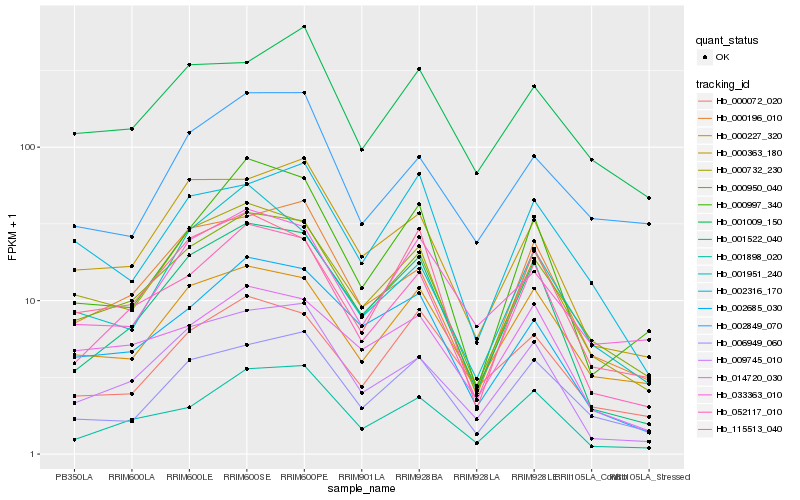
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000950_040 |
0.0 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 2 |
Hb_000732_230 |
0.0656814258 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_052117_010 |
0.082658644 |
- |
- |
kinase, putative [Ricinus communis] |
| 4 |
Hb_000227_320 |
0.0856036745 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_000196_010 |
0.0882264457 |
- |
- |
Patellin-5, putative [Ricinus communis] |
| 6 |
Hb_006949_060 |
0.0951708301 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 7 |
Hb_115513_040 |
0.0971574675 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002685_030 |
0.1014259129 |
- |
- |
PREDICTED: protein SMG7L [Jatropha curcas] |
| 9 |
Hb_000363_180 |
0.1038644956 |
- |
- |
PREDICTED: uncharacterized protein LOC105633052 [Jatropha curcas] |
| 10 |
Hb_009745_010 |
0.1066800575 |
- |
- |
cellulose synthase-like protein [Populus tomentosa] |
| 11 |
Hb_001522_040 |
0.1069746301 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 12 |
Hb_033363_010 |
0.1075798717 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 13 |
Hb_001898_020 |
0.1091166207 |
- |
- |
PREDICTED: uncharacterized protein LOC105629659 [Jatropha curcas] |
| 14 |
Hb_001009_150 |
0.1123273835 |
- |
- |
Glyceraldehyde-3-phosphate dehydrogenase [Gossypium arboreum] |
| 15 |
Hb_001951_240 |
0.1159747449 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |
| 16 |
Hb_014720_030 |
0.116523869 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0012s08280g [Populus trichocarpa] |
| 17 |
Hb_002849_070 |
0.1177839467 |
- |
- |
PREDICTED: protein BPS1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000072_020 |
0.1185532888 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000997_340 |
0.1223412828 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 20 |
Hb_002316_170 |
0.1226581712 |
- |
- |
purine permease, putative [Ricinus communis] |