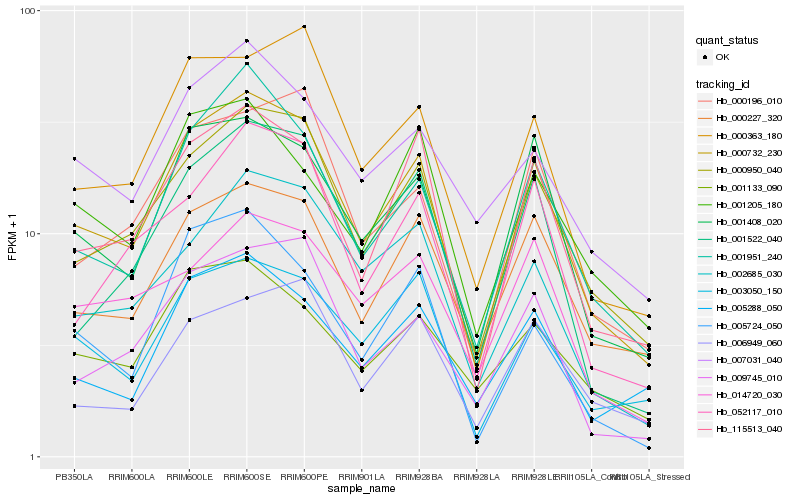
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000732_230 |
0.0 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000950_040 |
0.0656814258 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 3 |
Hb_115513_040 |
0.0834254814 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001408_020 |
0.0878955843 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 2-like [Jatropha curcas] |
| 5 |
Hb_001951_240 |
0.0929975928 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |
| 6 |
Hb_000227_320 |
0.0986694665 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_003050_150 |
0.1006505794 |
- |
- |
endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase, putative [Ricinus communis] |
| 8 |
Hb_000363_180 |
0.1035464601 |
- |
- |
PREDICTED: uncharacterized protein LOC105633052 [Jatropha curcas] |
| 9 |
Hb_000196_010 |
0.106304705 |
- |
- |
Patellin-5, putative [Ricinus communis] |
| 10 |
Hb_002685_030 |
0.1070377123 |
- |
- |
PREDICTED: protein SMG7L [Jatropha curcas] |
| 11 |
Hb_001522_040 |
0.1093675377 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 12 |
Hb_052117_010 |
0.1107210078 |
- |
- |
kinase, putative [Ricinus communis] |
| 13 |
Hb_009745_010 |
0.1133161977 |
- |
- |
cellulose synthase-like protein [Populus tomentosa] |
| 14 |
Hb_005288_050 |
0.1182833109 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR4 [Jatropha curcas] |
| 15 |
Hb_007031_040 |
0.1185952114 |
transcription factor |
TF Family: bZIP |
PREDICTED: bZIP transcription factor 17-like [Jatropha curcas] |
| 16 |
Hb_001205_180 |
0.1188768458 |
- |
- |
hypothetical protein POPTR_0005s12580g [Populus trichocarpa] |
| 17 |
Hb_001133_090 |
0.1191333946 |
- |
- |
hypothetical protein POPTR_0012s02680g [Populus trichocarpa] |
| 18 |
Hb_014720_030 |
0.1228303295 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0012s08280g [Populus trichocarpa] |
| 19 |
Hb_006949_060 |
0.1230662392 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 20 |
Hb_005724_050 |
0.1238281205 |
- |
- |
- |