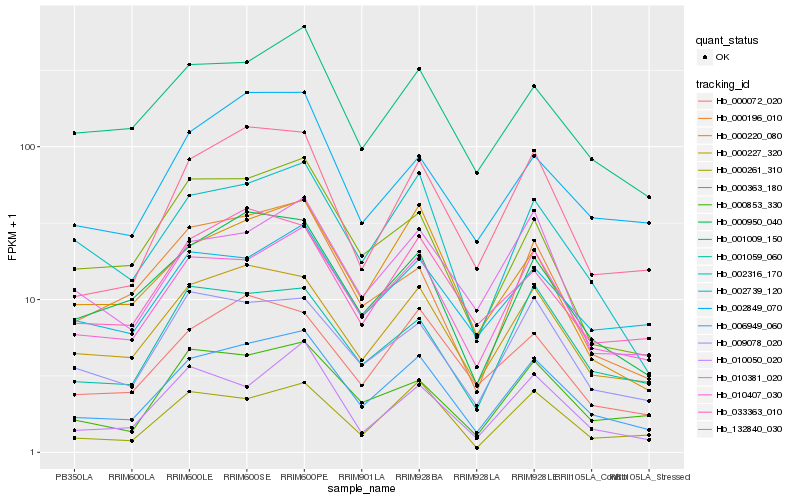
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006949_060 |
0.0 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 2 |
Hb_010407_030 |
0.06854752 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 3 |
Hb_000227_320 |
0.0861794346 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_132840_030 |
0.0943293851 |
transcription factor |
TF Family: MYB-related |
lhy1 [Populus tremula] |
| 5 |
Hb_000950_040 |
0.0951708301 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 6 |
Hb_001009_150 |
0.0995443354 |
- |
- |
Glyceraldehyde-3-phosphate dehydrogenase [Gossypium arboreum] |
| 7 |
Hb_002316_170 |
0.1001402128 |
- |
- |
purine permease, putative [Ricinus communis] |
| 8 |
Hb_000853_330 |
0.1032471649 |
- |
- |
PREDICTED: importin subunit alpha-like [Jatropha curcas] |
| 9 |
Hb_001059_060 |
0.1083049771 |
- |
- |
PREDICTED: bifunctional riboflavin kinase/FMN phosphatase [Jatropha curcas] |
| 10 |
Hb_009078_020 |
0.1102258222 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 11 |
Hb_000072_020 |
0.1131895348 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000196_010 |
0.1136829298 |
- |
- |
Patellin-5, putative [Ricinus communis] |
| 13 |
Hb_002739_120 |
0.1149193593 |
- |
- |
PREDICTED: zinc finger protein ZPR1-like [Jatropha curcas] |
| 14 |
Hb_000261_310 |
0.1163142926 |
- |
- |
PREDICTED: UV radiation resistance-associated gene protein isoform X3 [Jatropha curcas] |
| 15 |
Hb_002849_070 |
0.1165931708 |
- |
- |
PREDICTED: protein BPS1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000220_080 |
0.1172632177 |
- |
- |
kinase, putative [Ricinus communis] |
| 17 |
Hb_010381_020 |
0.1179575927 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 18 |
Hb_033363_010 |
0.1184275253 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 19 |
Hb_010050_020 |
0.1204210268 |
- |
- |
PREDICTED: uncharacterized protein LOC102601222 [Solanum tuberosum] |
| 20 |
Hb_000363_180 |
0.1215155943 |
- |
- |
PREDICTED: uncharacterized protein LOC105633052 [Jatropha curcas] |