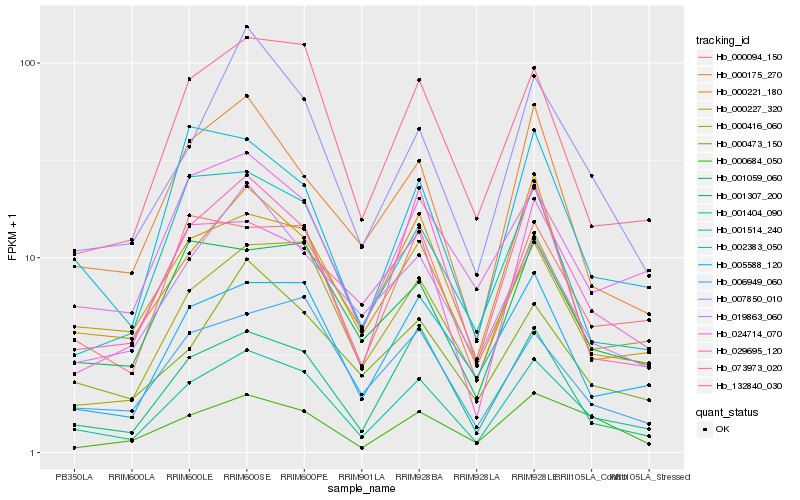
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001404_090 |
0.0 |
- |
- |
PREDICTED: kinesin-like protein NACK2 [Camelina sativa] |
| 2 |
Hb_000416_060 |
0.106626092 |
- |
- |
PREDICTED: uncharacterized protein LOC105649718 [Jatropha curcas] |
| 3 |
Hb_000094_150 |
0.1087956253 |
- |
- |
- |
| 4 |
Hb_132840_030 |
0.1110816676 |
transcription factor |
TF Family: MYB-related |
lhy1 [Populus tremula] |
| 5 |
Hb_005588_120 |
0.1141879695 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001307_200 |
0.1185390332 |
- |
- |
PREDICTED: uncharacterized protein LOC105644489 [Jatropha curcas] |
| 7 |
Hb_073973_020 |
0.1231849588 |
- |
- |
PREDICTED: DIS3-like exonuclease 2 [Jatropha curcas] |
| 8 |
Hb_024714_070 |
0.1253256306 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 9 |
Hb_001059_060 |
0.1253902644 |
- |
- |
PREDICTED: bifunctional riboflavin kinase/FMN phosphatase [Jatropha curcas] |
| 10 |
Hb_000227_320 |
0.1258287239 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_000221_180 |
0.1264967142 |
- |
- |
VQ motif-containing family protein [Populus trichocarpa] |
| 12 |
Hb_007850_010 |
0.1297839141 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.4 [Jatropha curcas] |
| 13 |
Hb_002383_050 |
0.1311329176 |
- |
- |
PREDICTED: uncharacterized protein LOC105628744 [Jatropha curcas] |
| 14 |
Hb_000175_270 |
0.1313040533 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001514_240 |
0.1330720784 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 16 |
Hb_029695_120 |
0.1330724785 |
- |
- |
hypothetical protein POPTR_0002s05320g [Populus trichocarpa] |
| 17 |
Hb_000473_150 |
0.1332994923 |
- |
- |
PREDICTED: pullulanase 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000684_050 |
0.1350644249 |
- |
- |
PREDICTED: MLO-like protein 6 [Sesamum indicum] |
| 19 |
Hb_019863_060 |
0.1353006167 |
- |
- |
PREDICTED: target of Myb protein 1-like [Populus euphratica] |
| 20 |
Hb_006949_060 |
0.1372112846 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |