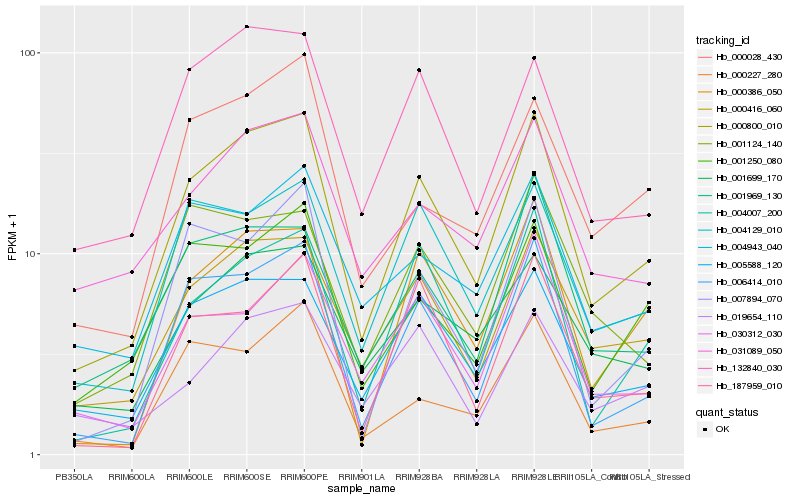
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000800_010 |
0.0 |
- |
- |
PREDICTED: glycosyltransferase family protein 64 protein C5 [Jatropha curcas] |
| 2 |
Hb_000416_060 |
0.0893444465 |
- |
- |
PREDICTED: uncharacterized protein LOC105649718 [Jatropha curcas] |
| 3 |
Hb_005588_120 |
0.1020193105 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_006414_010 |
0.1032811062 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 9 [Jatropha curcas] |
| 5 |
Hb_004129_010 |
0.1172448236 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 6 |
Hb_001699_170 |
0.1193451978 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_132840_030 |
0.1202435269 |
transcription factor |
TF Family: MYB-related |
lhy1 [Populus tremula] |
| 8 |
Hb_004007_200 |
0.1207501676 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY5 [Jatropha curcas] |
| 9 |
Hb_000386_050 |
0.1231763496 |
- |
- |
PREDICTED: 2-isopropylmalate synthase 2, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_001969_130 |
0.1240235226 |
- |
- |
PREDICTED: molybdate transporter 2 [Jatropha curcas] |
| 11 |
Hb_001250_080 |
0.1249343854 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g23390 [Jatropha curcas] |
| 12 |
Hb_031089_050 |
0.1275029443 |
- |
- |
kinase, putative [Ricinus communis] |
| 13 |
Hb_007894_070 |
0.1344415795 |
- |
- |
PREDICTED: uncharacterized protein LOC105632955 [Jatropha curcas] |
| 14 |
Hb_030312_030 |
0.1354078618 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 15 |
Hb_187959_010 |
0.1359221475 |
- |
- |
hypothetical protein JCGZ_12266 [Jatropha curcas] |
| 16 |
Hb_000028_430 |
0.1362196478 |
- |
- |
PREDICTED: uncharacterized protein LOC105641369 isoform X1 [Jatropha curcas] |
| 17 |
Hb_004943_040 |
0.1365264664 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g30440 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000227_280 |
0.1366291639 |
- |
- |
PREDICTED: putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g14440 [Jatropha curcas] |
| 19 |
Hb_001124_140 |
0.1371759475 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |
| 20 |
Hb_019654_110 |
0.1390465508 |
- |
- |
PREDICTED: F-box protein SKIP23-like [Jatropha curcas] |