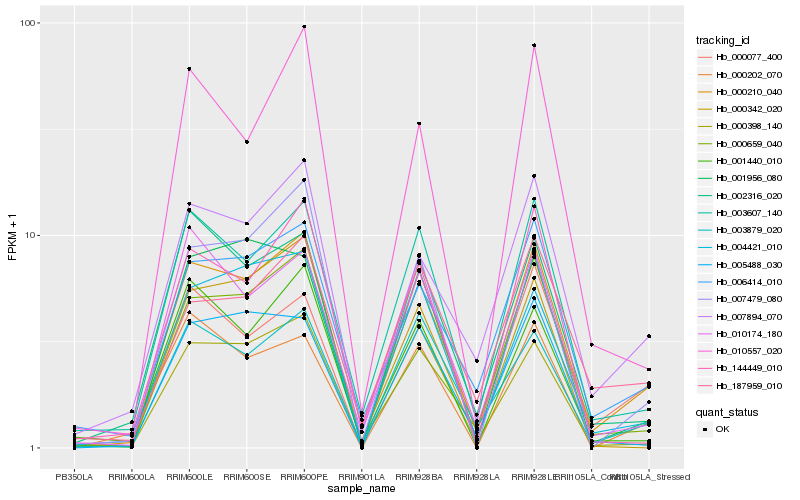
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000210_040 |
0.0 |
- |
- |
PREDICTED: putative HVA22-like protein g [Jatropha curcas] |
| 2 |
Hb_007894_070 |
0.1000135659 |
- |
- |
PREDICTED: uncharacterized protein LOC105632955 [Jatropha curcas] |
| 3 |
Hb_003607_140 |
0.1035653701 |
- |
- |
3-ketoacyl-CoA thiolase B, putative [Ricinus communis] |
| 4 |
Hb_006414_010 |
0.1067523976 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 9 [Jatropha curcas] |
| 5 |
Hb_004421_010 |
0.1175665043 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 6 |
Hb_003879_020 |
0.1185427603 |
- |
- |
PREDICTED: MATE efflux family protein 1-like [Jatropha curcas] |
| 7 |
Hb_000659_040 |
0.1226060037 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_002316_020 |
0.125195965 |
- |
- |
PREDICTED: uncharacterized protein LOC105640407 [Jatropha curcas] |
| 9 |
Hb_010174_180 |
0.1291586952 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_000398_140 |
0.1322762064 |
- |
- |
PREDICTED: methionine S-methyltransferase [Jatropha curcas] |
| 11 |
Hb_010557_020 |
0.1328091148 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 12 |
Hb_001956_080 |
0.1350408892 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 14 [Jatropha curcas] |
| 13 |
Hb_007479_080 |
0.1374531727 |
- |
- |
PREDICTED: uncharacterized protein LOC105649774 isoform X1 [Jatropha curcas] |
| 14 |
Hb_005488_030 |
0.1384217082 |
- |
- |
F3F9.11 [Arabidopsis lyrata subsp. lyrata] |
| 15 |
Hb_144449_010 |
0.1395382302 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 16 |
Hb_001440_010 |
0.1407128332 |
- |
- |
PREDICTED: armadillo repeat-containing kinesin-like protein 1 [Populus euphratica] |
| 17 |
Hb_000202_070 |
0.1435265844 |
- |
- |
putative RNA-directed DNA polymerase (Reverse transcriptase) [Malus domestica] |
| 18 |
Hb_187959_010 |
0.1456184865 |
- |
- |
hypothetical protein JCGZ_12266 [Jatropha curcas] |
| 19 |
Hb_000077_400 |
0.1459958808 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_000342_020 |
0.146206944 |
- |
- |
bel1 homeotic protein, putative [Ricinus communis] |