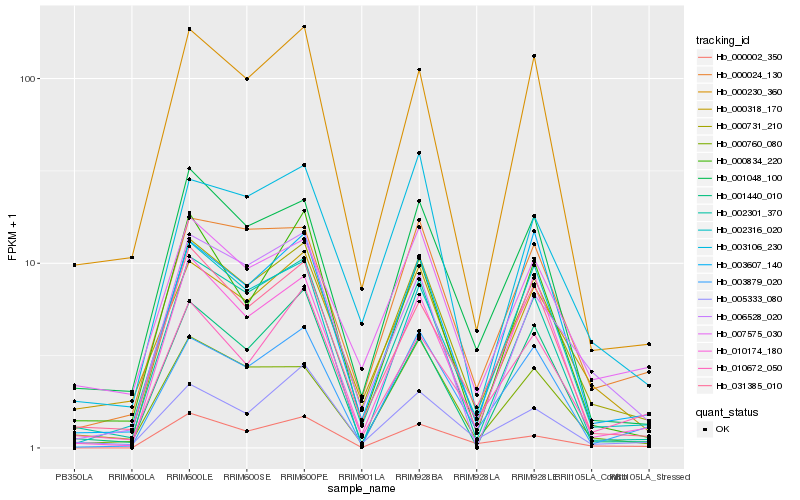
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000731_210 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002301_370 |
0.0891607402 |
- |
- |
hypothetical protein JCGZ_21454 [Jatropha curcas] |
| 3 |
Hb_000024_130 |
0.0960557123 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001048_100 |
0.1026910402 |
- |
- |
PREDICTED: lysosomal Pro-X carboxypeptidase [Jatropha curcas] |
| 5 |
Hb_031385_010 |
0.1028057206 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 6 |
Hb_000318_170 |
0.104790875 |
- |
- |
N-acetyl-gamma-glutamyl-phosphate reductase, putative [Ricinus communis] |
| 7 |
Hb_003106_230 |
0.108887608 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM1-like [Jatropha curcas] |
| 8 |
Hb_010174_180 |
0.1089818891 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 9 |
Hb_003879_020 |
0.1120152441 |
- |
- |
PREDICTED: MATE efflux family protein 1-like [Jatropha curcas] |
| 10 |
Hb_000760_080 |
0.1128587681 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 11 |
Hb_003607_140 |
0.1153844451 |
- |
- |
3-ketoacyl-CoA thiolase B, putative [Ricinus communis] |
| 12 |
Hb_000230_360 |
0.1154511687 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 13 |
Hb_002316_020 |
0.1190934272 |
- |
- |
PREDICTED: uncharacterized protein LOC105640407 [Jatropha curcas] |
| 14 |
Hb_006528_020 |
0.1195996606 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_007575_030 |
0.1215306846 |
- |
- |
PREDICTED: protein RCC2 homolog [Jatropha curcas] |
| 16 |
Hb_005333_080 |
0.1220664114 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_010672_050 |
0.1240375544 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 isoform X3 [Jatropha curcas] |
| 18 |
Hb_000002_350 |
0.1246897994 |
transcription factor |
TF Family: SNF2 |
DNA repair helicase rad5,16, putative [Ricinus communis] |
| 19 |
Hb_000834_220 |
0.1267330253 |
- |
- |
PREDICTED: uncharacterized protein LOC105637705 [Jatropha curcas] |
| 20 |
Hb_001440_010 |
0.1281919163 |
- |
- |
PREDICTED: armadillo repeat-containing kinesin-like protein 1 [Populus euphratica] |