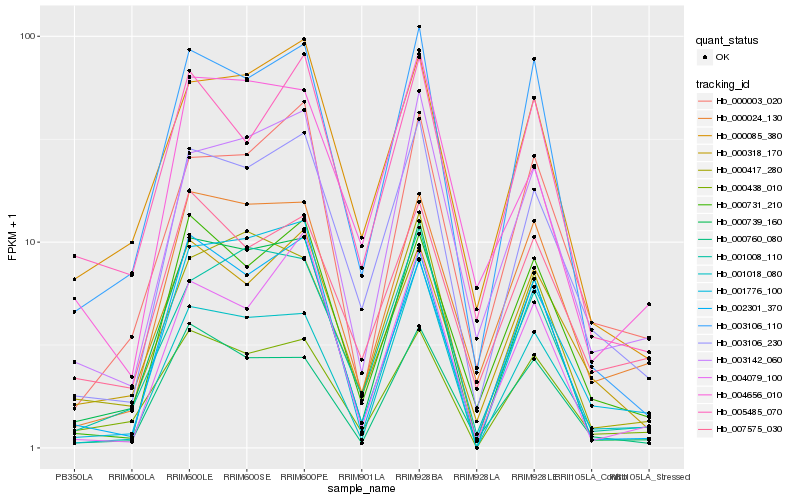
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003106_230 |
0.0 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM1-like [Jatropha curcas] |
| 2 |
Hb_003142_060 |
0.1041746195 |
- |
- |
PREDICTED: glutamate synthase 1 [NADH], chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_000731_210 |
0.108887608 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000318_170 |
0.1145047944 |
- |
- |
N-acetyl-gamma-glutamyl-phosphate reductase, putative [Ricinus communis] |
| 5 |
Hb_001008_110 |
0.1158595376 |
- |
- |
- |
| 6 |
Hb_000024_130 |
0.1178115859 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000085_380 |
0.1184927534 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003106_110 |
0.1206148208 |
- |
- |
betaine aldehyde dehydrogenase 1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000003_020 |
0.1217103407 |
- |
- |
PREDICTED: uncharacterized protein LOC105631161 [Jatropha curcas] |
| 10 |
Hb_000739_160 |
0.1249425437 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_001018_080 |
0.1263267616 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004656_010 |
0.1277133015 |
- |
- |
PREDICTED: probable calcium-binding protein CML49 [Jatropha curcas] |
| 13 |
Hb_004079_100 |
0.1293743247 |
- |
- |
PREDICTED: uncharacterized protein LOC105637705 [Jatropha curcas] |
| 14 |
Hb_000438_010 |
0.1308115863 |
- |
- |
- |
| 15 |
Hb_000417_280 |
0.1319054574 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_007575_030 |
0.1333900548 |
- |
- |
PREDICTED: protein RCC2 homolog [Jatropha curcas] |
| 17 |
Hb_000760_080 |
0.1357920527 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 18 |
Hb_005485_070 |
0.1365142272 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |
| 19 |
Hb_002301_370 |
0.1391439389 |
- |
- |
hypothetical protein JCGZ_21454 [Jatropha curcas] |
| 20 |
Hb_001776_100 |
0.1394055661 |
- |
- |
PREDICTED: outer envelope pore protein 16, chloroplastic [Jatropha curcas] |