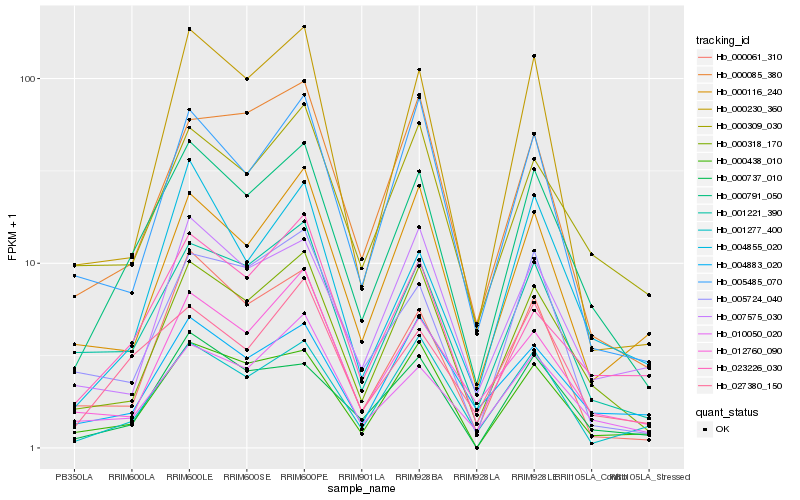
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000791_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636760 [Jatropha curcas] |
| 2 |
Hb_000318_170 |
0.0960866923 |
- |
- |
N-acetyl-gamma-glutamyl-phosphate reductase, putative [Ricinus communis] |
| 3 |
Hb_001221_390 |
0.1115016222 |
- |
- |
Phospholipase C 4 precursor, putative [Ricinus communis] |
| 4 |
Hb_027380_150 |
0.113140573 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000737_010 |
0.1257268965 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RING1a isoform X1 [Jatropha curcas] |
| 6 |
Hb_010050_020 |
0.1263543948 |
- |
- |
PREDICTED: uncharacterized protein LOC102601222 [Solanum tuberosum] |
| 7 |
Hb_000438_010 |
0.129309729 |
- |
- |
- |
| 8 |
Hb_000309_030 |
0.13097386 |
- |
- |
PREDICTED: formate dehydrogenase, mitochondrial [Jatropha curcas] |
| 9 |
Hb_000061_310 |
0.136198065 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |
| 10 |
Hb_023226_030 |
0.1384125585 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-14-like [Jatropha curcas] |
| 11 |
Hb_001277_400 |
0.1401397373 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 4B isoform X3 [Jatropha curcas] |
| 12 |
Hb_000230_360 |
0.1405395872 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 13 |
Hb_005485_070 |
0.143307582 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |
| 14 |
Hb_004855_020 |
0.1441375581 |
- |
- |
hypothetical protein POPTR_0006s10010g [Populus trichocarpa] |
| 15 |
Hb_012760_090 |
0.1442525747 |
- |
- |
Aspartic proteinase-like protein 1 [Morus notabilis] |
| 16 |
Hb_000116_240 |
0.1450821069 |
transcription factor |
TF Family: HB |
homeobox-leucine zipper family protein [Populus trichocarpa] |
| 17 |
Hb_007575_030 |
0.1456262092 |
- |
- |
PREDICTED: protein RCC2 homolog [Jatropha curcas] |
| 18 |
Hb_004883_020 |
0.1457602286 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 19 |
Hb_005724_040 |
0.1458250163 |
- |
- |
hypothetical protein JCGZ_13142 [Jatropha curcas] |
| 20 |
Hb_000085_380 |
0.149226543 |
- |
- |
conserved hypothetical protein [Ricinus communis] |